Oral
Machine Learning for Quantitative Imaging
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 16:00 - 18:00 | Moderators: Anahita Fathi Kazerooni & Frank Zöllner |
 |
0327.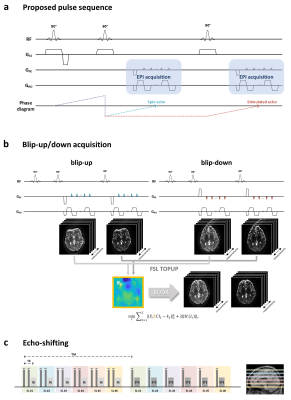 |
BUDA-STEAM: A rapid parameter estimation method for T1, T2, M0, B0 and B1 using three-90° pulse sequence
Seohee So1, Byungjai Kim1, HyunWook Park1, and Berkin Bilgic2
1Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Martinos Center for Biomedical Imaging, Charlestown, MA, United States
An MR parameter estimation method using spin- and stimulated-echo signals is proposed. Three-90° pulse sequence is introduced to simultaneously acquire spin and stimulated echo signals. These signals contain rich information that allow for estimation of T1, T2, M0, B0 and B1 maps. We utilize Blip Up-Down Acquisition (BUDA) to eliminate geometric distortion incurred by rapid EPI readout. In order to estimate the parameter maps from the spin- and stimulated-echo signals with high fidelity, two parameter estimation methods, analytic fitting and a novel unsupervised deep neural network method, are developed.
|
|
 |
0328.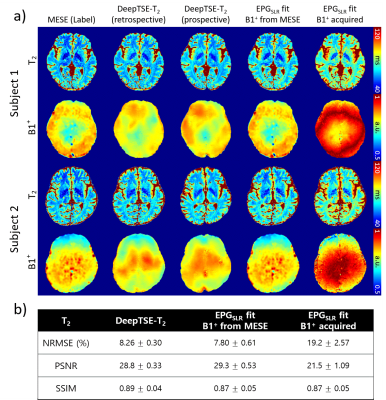 |
DeepTSE-T2: Deep learning-powered T2 mapping with B1+ estimation using a product double-echo Turbo Spin Echo sequence
Hwihun Jeong1, Hyeong-Geol Shin1, Sooyeon Ji1, Jinhee Jang2, Hyun-Soo Lee3, Yoonho Nam4, and Jongho Lee1
1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2Department of Radiology, Seoul St Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea, Republic of, 3Siemens healthineers Ltd, Seoul, Korea, Republic of, 4Division of Biomedical Engineering, Hankuk University of Foreign Studies, Yongin, Korea, Republic of
We developed DeepTSE-T2, a deep learning-based T2 mapping algorithm with retrospective B1+ estimation for a product double-echo TSE sequence. DeepTSE-T2 enables T2 mapping by retrospectively estimating B1+ information, reconstructing T2 in high-accuracy (NRMSE = 8.26 ± 0.30%). The proposed method is useful in a clinical setting since it utilizes a fast imaging product sequence. The training dataset consists of simulation-based data, providing flexibility in parameter setting. Applications to χ-separation and an MS patient are included.
|
|
 |
0329.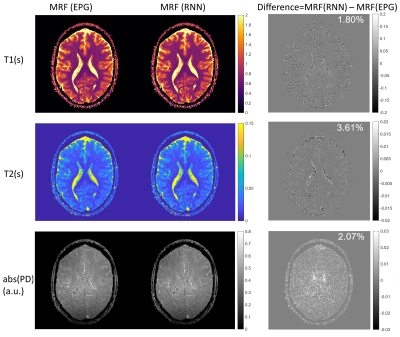 |
Fast and Accurate Modeling of Transient-state Sequences by Recurrent Neural Networks
Hongyan Liu1, Oscar van der Heide1, Cornelis A.T. van den Berg1, and Alessandro Sbrizzi1
1Computational Imaging Group for MR diagnostics & therapy, Center for Image Sciences, UMC Utrecht, Utrecht, Netherlands Fast and accurate modeling of transient-state sequences are required for various quantitative MR applications. We present here a surrogate model based on Recurrent Neural Network (RNN) architecture, to quickly compute large-scale MR signals and derivatives. We demonstrate that the trained RNN model works with different sequence parameters and tiussue parameters without the need of retraining. We prove that the RNN model can be used for computing large-scale MR signals and derivatives within seconds, and therefore achieves one to three orders of magnitude acceleration for different qMRI applications. |
|
0330.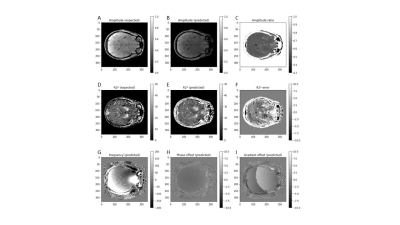 |
Unsupervised physics-informed deep learning (N=1) for solving inverse qMRI problems – Relaxometry and field mapping from multi-echo data
Ilyes Benslimane1, Thomas Jochmann2, Robert Zivadinov1,3, and Ferdinand Schweser1,3
1Buffalo Neuroimaging Analysis Center, Department of Neurology, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, The State University of New York, Buffalo, NY, United States, Buffalo, NY, United States, 2Department of Computer Science and Automation, Technische Universität Ilmenau, Ilmenau, Germany, Jena, Thuringia, Germany, 3Center for Biomedical Imaging, Clinical and Translational Science Institute at the University at Buffalo, Buffalo, NY, USA, Buffalo, NY, United States
Modeling the non-linear relationship of the Magnetic Resonance (MR) signal and biophysical sources is computationally expensive and unstable using conventional methods. We develop an unsupervised physics-informed deep learning algorithm that quantifies MR parameters from multi-echo GRE data in a single computational pass. The algorithm produced accurate B0 and R2* field maps without phase wrapping artifacts and with typical contrast variations. The success of this network demonstrates the feasibility of physics-informed quantitative MRI (qMRI) without the need for ground truth training data, typically required by similar networks. This developed tool could provide fast and comprehensive tissue characterization in qMRI.
|
||
0331.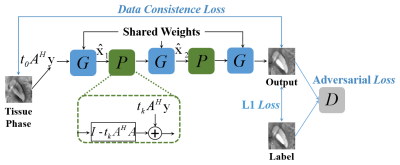 |
MoG-QSM: A Model-based Generative Adversarial Deep Learning Network for Quantitative Susceptibility Mapping
Ruimin Feng1, Yuting Shi1, Jie Feng1, Yuyao Zhang2, and Hongjiang Wei1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2School of Information Science and Technology, ShanghaiTech University, Shanghai, China
We proposed a model-based generative adversarial network for quantitative susceptibility mapping. Total 30 scans from six healthy subjects, acquired at five different head orientations, were employed for network training. The trained network provided superior image quality and accuracy quantification compared to recently developed QSM reconstruction methods. The proposed method showed excellent tissue susceptibility contrast and artifact suppression on the QSM images of patients with hemorrhage and multiple sclerosis, demonstrating potential clinical application in the future.
|
||
0332.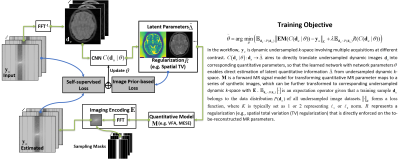 |
Self-supervised Deep Learning for Rapid Quantitative Imaging
Fang Liu1 and Li Feng2
1Radiology, Harvard Medical School, Boston, MA, United States, 2Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States
The purpose of this work was to develop a model-guided self-supervised deep learning MRI reconstruction framework called REference-free LAtent map eXtraction (RELAX) for rapid quantitative MR parameter mapping. RELAX eliminates the need for full sampled reference datasets that are required in standard supervised learning. Meanwhile, RELAX also enables direct reconstruction of MR parameter maps from undersampled k-space. Our results demonstrated that the proposed framework produced accurate and robust T1/T2 mapping in accelerated and low-SNR MRI. The good quantitative agreement to the reference method suggests that RELAX allows accelerated quantitative imaging without training with reference data.
|
||
0333.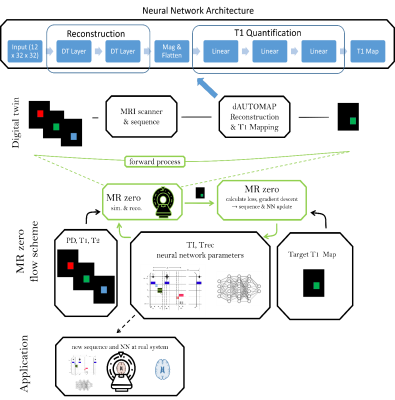 |
MRzero with dAUTOMAP reconstruction– automated invention of MR acquisition and neural network reconstruction
Hoai Nam Dang1, Simon Weinmüller1, Alexander Loktyushin2,3, Felix Glang2, Arnd Dörfler1, Andreas Maier4, Bernhard Schölkopf3, Klaus Scheffler2,5, and Moritz Zaiss1,2
1Neuroradiology, University Clinic Erlangen, Friedrich-Alexander Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 2Magnetic Resonance Center, Max-Planck Institute for Biological Cybernetics, Tübingen, Germany, 3Empirical Inference, Max-Planck Institute for Intelligent Systems, Tübingen, Germany, 4Pattern Recognition Lab, Friedrich-Alexander Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 5Department of Biomedical Magnetic Resonance, Eberhard Karls University Tübingen, Tübingen, Germany
We present an end-to-end optimized T1 mapping utilizing MRzero - a fully differentiable Bloch-equation-based MRI sequence invention framework. A convolutional neural network is employed for combined image reconstruction and parameter mapping. The pipeline performs a joint optimization of sequence parameters and neural network parameters to create a full autoencoder for T1 mapping. We demonstrate for in vivo measurements at 3T, that the CNN based reconstruction and T1 mapping outperformes a conventional reconstruction with pixelwise neural network based T1 quantification.
|
||
0334.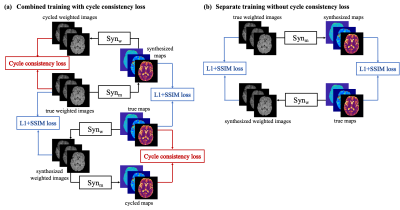 |
Bidirectional Translation Between Multi-Contrast Images and Multi-Parametric Maps Using Deep Learning
Shihan Qiu1,2, Yuhua Chen1,2, Sen Ma1, Zhaoyang Fan1,2, Anthony G. Christodoulou1,2, Yibin Xie1, and Debiao Li1,2
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, UCLA, Los Angeles, CA, United States
Multi-contrast MRI and multi-parametric maps provide complementary qualitative and quantitative information for disease diagnosis. However, due to limited scan time, a full array of images is often unavailable in practice. To provide both qualitative weighted images and quantitative maps using a weighted-only or mapping-only acquisition, in this work, we propose to perform bidirectional translation between conventional weighted images and parametric maps. We developed a combined training strategy of two convolutional neural networks with cycle consistency loss. Our preliminary results show that the proposed method can translate between contrast-weighted images and quantitative maps with high quality and fidelity.
|
||
0335.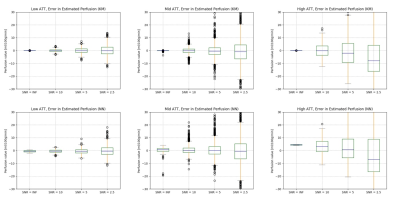 |
Accelerating perfusion quantification using ASL-MRI with a neural network based forward model
Yechuan Zhang1 and Michael A Chappell1,2,3
1Institute of Biomedical Engineering, Department of Engineering Science, University of Oxford, Oxford, United Kingdom, 2Sir Peter Mansfield Imaging Centre, School of Medicine, University of Nottingham, Nottingham, United Kingdom, 3Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
Arterial Spin Labelling is established as a quantitative technique to measure perfusion and other hemodynamic properties of the cerebral vasculature. This application of ASL requires multiple post label delays and parameter estimation via a kinetic model. However, the computational cost of the post-processing can be an issue, especially with sophisticated kinetic models. In this work, we propose a rapid method to perform perfusion estimation by replacing kinetic models with pre-trained neural networks. Two neural networks were trained to replace the kinetic model with or without gamma dispersion effects. The dispersion neural network is shown to achieve a lower computational cost.
|
||
0336.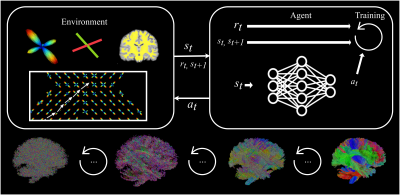 |
Track-To-Learn: A general framework for tractography with deep reinforcement learning
Antoine Théberge1, Christian Desrosiers2, Maxime Descoteaux1, and Pierre-Marc Jodoin1
1Faculté des Sciences, Université de Sherbrooke, Sherbrooke, QC, Canada, 2Département de génie logiciel et des TI, École de technologie supérieure, Montréal, QC, Canada
Supervised machine learning algorithms have been proposed to learn tractography algorithms implicitly from data, without relying on hard-to-develop anatomical priors. However, supervised learning methods rely on labelled data that is very hard to obtain. To remove the need for such data but still leverage the expressiveness of neural networks, we introduce and implement Track-To-Learn, a general framework to pose tractography as a deep reinforcement learning problem. We show that competitive results can be obtained on known data and that the learned algorithms are able to generalize far better to new, unseen data, than prior supervised learning-based tractography algorithms.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.