Digital Poster Session
Young Investigator Award Posters
Session Topic: Young Investigator Awards
Session Sub-Topic: Young Investigator Awards
YIA Poster
Other
0001.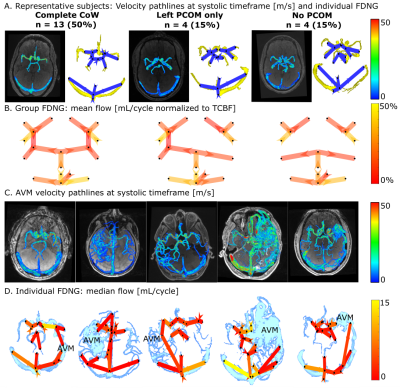 |
Standardized Evaluation of Cerebral Arteriovenous Malformations using Flow Distribution Network Graphs and Dual-venc 4D Flow MRI
Maria Aristova1, Alireza Vali1, Sameer A Ansari1,2,3, Ali Shaibani1,2, Tord D Alden2,4, Michael C Hurley1,2, Babak S Jahromi1,2, Matthew B Potts1,2, Michael Markl1,5, and Susanne Schnell1
1Radiology, Northwestern University, Chicago, IL, United States, 2Department of Neurosurgery, Northwestern University, Chicago, IL, United States, 3Department of Neurology, Northwestern University, Chicago, IL, United States, 4Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, United States, 5McCormick School of Engineering, Biomedical Engineering, Northwestern University, Chicago, IL, United States
Dual-venc 4D flow MRI with PEAK-GRAPPA acceleration provides time-resolved 3D cerebral hemodynamics and could be applied to cerebral arteriovenous malformations (AVM) with an appropriate standardized protocol. We optimize dual-venc 4D flow imaging for AVM in vitro and in vivo, and apply a Flow Distribution Network Graph paradigm for storing and analyzing complex neurovascular 4D flow data. In vitro and in vivo, 4 voxels across a typical vessel (achievable in vivo with 0.8mm isotropic resolution) will yield flow conservation < 15% and high reproducibility. Venous-arterial ratios of peak velocity and pulsatility index are proposed as potential network-based biomarkers characterizing AVM hemodynamics.
|
|
| 0002. | Parametric Hemodynamic 4D flow MRI maps for the Characterization of Chronic Thoracic Descending Aortic Dissection
Kelly Jarvis1, Judith T Pruijssen2, Andre Y Son3, Bradley D Allen1, Gilles Soulat1, Alireza Vali1, Alex J Barker4, Andrew W Hoel5, Mark K Eskandari5, S. Chris Malaisrie3, James C Carr1, Jeremy D Collins6, and Michael Markl1
1Department of Radiology, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States, 2Department of Radiology and Nuclear Medicine, Radboud University Medical Centre, Nijmegen, Netherlands, 3Division of Cardiac Surgery, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States, 4Department of Radiology, University of Colorado, Denver, CO, United States, 5Division of Vascular Surgery, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States, 66. Department of Radiology, Mayo Clinic, Rochester, MN, United States
Systematic evaluation of complex flow in descending aortic dissection (DAD) is needed to better understand which patients are predisposed to complications. Our goal was to utilize quantitative maps from 4D flow MRI for monitoring true and false lumen (TL, FL) flow characteristics. 4D flow was acquired in 20 DAD patients (6 medically managed, 14 with surgical repair), and 21 age-matched controls. 4D flow-derived quantitative maps demonstrated global and regional hemodynamic differences between DAD patients and controls. DAD patients with and without repair showed significantly altered TL and FL aortic hemodynamics, indicating this technique’s potential to characterize flow dynamics in DAD.
|
|
0003.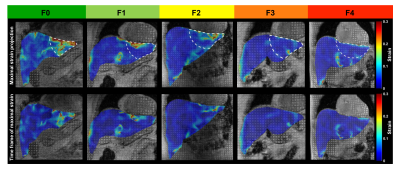 |
MRI Cine-Tagging of Cardiac-Induced Motion for Noninvasive Staging of Liver Fibrosis
Thierry Lefebvre1,2,3, Léonie Petitclerc1,2,4, Mélanie Hébert1,2, Laurent Bilodeau1,2, Giada Sebastiani5, Damien Olivié1, Zu-Hua Gao6, Marie-Pierre Sylvestre2,7, Guy Cloutier1,8,9, Bich N Nguyen10, Guillaume Gilbert1,11, and An Tang1,2,8
1Radiology, Radio-Oncology and Nuclear Medicine, Université de Montréal, Montreal, QC, Canada, 2Centre de recherche du Centre hospitalier de l'Université de Montréal (CRCHUM), Montreal, QC, Canada, 3Medical Physics Unit, McGill University, Montréal, QC, Canada, 4C.J. Gorter Center for High Field MRI, Department of Radiology, Leiden University Medical Center (LUMC), Leiden, Netherlands, 5Department of Medicine, Division of Gastroenterology and Hepatology, McGill University Health Centre (MUHC), Montreal, QC, Canada, 6Department of Pathology, McGill University, Montreal, QC, Canada, 7Department of Social and Preventive Medicine, École de santé publique de l’Université de Montréal (ESPUM), Montreal, QC, Canada, 8Institute of Biomedical Engineering, Université de Montréal, Montreal, QC, Canada, 9Laboratory of Biorheology and Medical Ultrasonics (LBUM), Centre de recherche du Centre hospitalier de l’Université de Montréal (CRCHUM), Montreal, QC, Canada, 10Service of Pathology, Centre hospitalier de l'Université de Montréal (CHUM), Montreal, QC, Canada, 11MR Clinical Science, Philips Healthcare Canada, Montreal, QC, Canada
MR elastography techniques for staging liver fibrosis assess the right liver and require additional hardware. MRI cine-tagging evaluates the strain of liver tissue and shows promise for staging liver fibrosis without additional hardware. It can be performed routinely during MRI examinations. Strain showed high correlation with fibrosis stages (ρ = -0.68, P < 0.0001). AUC was 0.81 to distinguish fibrosis stages F0 vs. ≥F1, 0.84 for ≤F1 vs. ≥F2, 0.86 for ≤F2 vs. ≥F3, and 0.87 for ≤F3 vs. F4. It could be used to assess the left liver lobe as a complement to MR elastography assessing the right lobe.
|
|
0004.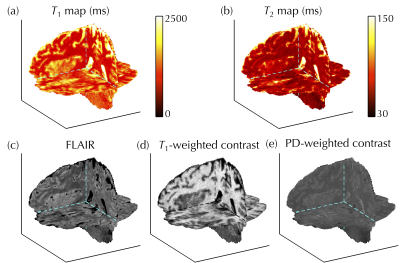 |
Multi-pathway multi-echo acquisition and contrast translation to generate a variety of quantitative and qualitative image contrasts
Cheng-Chieh Cheng1,2, Frank Preiswerk1, and Bruno Madore1
1Department of Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States, 2Department of Computer Science and Engineering, National Sun Yat-sen University, Kaohsiung, Taiwan
Ideally, neuro exams would include a variety of contrast types along with basic MRI parametric maps, with full-brain 3D coverage and good spatial resolution. However, tradeoffs exist between the number of contrasts, spatial coverage, spatial resolution, and scan time. We developed a 3D multi-pathway multi-echo (MPME) sequence that rapidly captures vast amounts of information about the object, and a ‘contrast translator’ to convert this information into desired contrasts. More specifically, a neural network converts 3D full-brain MPME data acquired in about 7 min into MPRAGE, FLAIR, T1W, T2W, T1 and T2 volumes, with the goal of abbreviating neuro exams.
|
|
0005.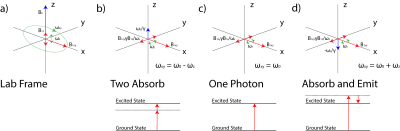 |
Multiphoton Magnetic Resonance Imaging
Victor Han1 and Chunlei Liu1,2
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2Helen Wills Neuroscience Institute, University of California, Berkeley, Berkeley, CA, United States
We present a fully geometric view of multiphoton excitation by taking a particular rotating frame transformation. In this rotating frame, we find that multiphoton excitations appear just like single-photon excitations again, and thus, we can readily generalize concepts already explored in standard single-photon excitation. With a homebuilt low-frequency (~ kHz) coil, we execute a standard slice-selective pulse sequence with all of its excitations replaced by their equivalent two-photon versions. With a multiphoton interpretation of oscillating gradients, we present a novel way to transform a standard slice-selective adiabatic pulse into a multiband one without modifying the RF pulse shape itself.
|
|
0006.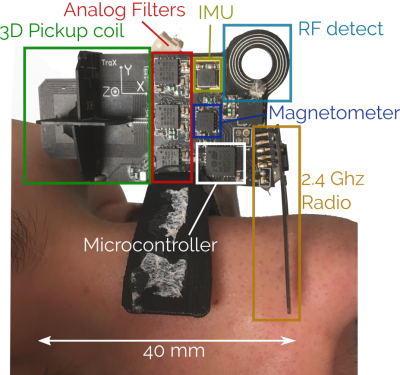 |
Toward “plug and play” prospective motion correction for MRI by combining observations of the time varying gradient and static vector fields.
Adam Marthinus Johannes van Niekerk1, Andre van der Kouwe1,2,3, and Ernesta Meintjes1,4,5
1Biomedical Engineering Research Centre, Division of Biomedical Engineering, Department of Human Biology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa, 2Athinoula A. Martinos Center, Massachusetts General Hospital, Charlestown, MA, United States, 3Radiology, Harvard Medical School, Boston, MA, United States, 4Cape Universities Body Imaging Centre, Cape Town, South Africa, 5Neuroscience Institute, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
Introducing additional hardware to measure patient motion allows for fast and accurate prospective motion correction that has minimal or no impact on the imaging pulse sequence. This does however entail additional setup that in some cases may be challenging to translate into a dynamic clinical setting. In this work we explore the use of an intelligent marker - a Wireless Radiofrequency-triggered Acquisition Device (WRAD) - for prospective motion correction. This new approach incorporates all additional hardware (besides a wireless receiver) into the marker that is attached to the subject. Initial results show improved image quality without scanner specific calibration.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Poster Video
Watch the Poster Video Back to Top
Back to Top