Digital Poster Session
Acquisition, Reconstruction & Analysis: Quantitative MRI
Acquisition, Reconstruction & Analysis
3736 -3751 Quantitative MRI - MR Fingerprinting 1
3752 -3767 Quantitative MRI - MR Fingerprinting 2
3768 -3783 Quantitative MRI - Quantitative Multi-Parameter Mapping
3784 -3799 Quantitative MRI - Quantitative MRI 1
3800 -3814 Quantitative MRI - Quantitative MRI 2
3736.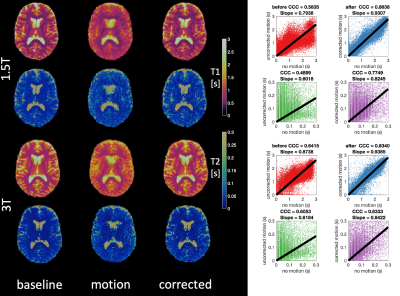 |
Retrospective motion correction of three-dimensional Magnetic Resonance Fingerprinting (MRF)
Jan W Kurzawski1,2, Matteo Cencini1,3, Luca Peretti1,3, Pedro A Gomez4, Rolf Schulte5, Graziella Donatelli1,6, Mirco Cosottini1,3, Paolo Cecchi6, Mauro Costagli1,7, Alessandra Retico2, Michela Tosetti1,7, and Guido Buonincontri1,7
1Imago7, Pisa, Italy, 2INFN, Pisa, Italy, 3University of Pisa, Pisa, Italy, 4Technical University of Munich, Munich, Germany, 5GE Healthcare, Munich, Germany, 6Neuroradiology, Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy, 7IRCCS Stella Maris, Pisa, Italy
We demonstrate a novel motion correction of 3D magnetic resonance fingerprinting (MRF) using spiral projection k-space trajectory. The motion was corrected using rigid motion parameters extracted from a whole-brain navigator obtained every 7s of imaging. Firstly, we optimized the trajectory ordering in simulation and selected the acquisition scheme that allowed the best navigator for motion correction. Secondly, we applied this scheme to invivo data in healthy subjects scanned first without motion and then while performing a motion paradigm. Our motion correction improved the correlation of motion-corrupted data with motionless data by over 20% for both T1 and T2.
|
|
3737.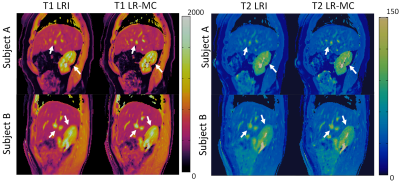 |
Generalised Low-Rank Non-rigid Motion Corrected reconstruction for 3D free breathing Liver Magnetic Resonance Fingerprinting
Gastao Cruz1, Olivier Jaubert1, Haikun Qi1, Torben Schneider2, René M. Botnar1, and Claudia Prieto1
1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Philips Healthcare, Guildford, United Kingdom
Magnetic Resonance Fingerprinting (MRF) has been shown to enable simultaneous T1 and T2 mapping of the liver and abdomen. 2D liver MRF requires breath holding, whereas preliminary results have been demonstrated for 3D free-breathing liver MRF using respiratory gating. However, gating approaches lead to unpredictable scan times and may impair the MRF encoding, since only data within the respiratory gating window is used for reconstruction. Here we propose a novel low-rank motion corrected approach to both resolve MRF varying contrast and perform non-rigid respiratory motion correction directly in the reconstruction, enabling 3D free-breathing liver MRF with 100% respiratory scan efficiency.
|
|
3738.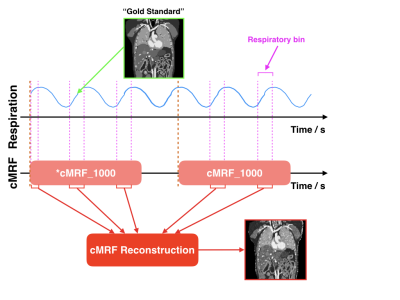 |
Motion Tracking using Continuous Magnetic Resonance Fingerprinting
Edward S. Hui1, Di Cui1, Jing Cai2, Queenie Chan3, and Peng Cao1
1Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong, 2Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 3Philips Healthcare, Hong Kong, Hong Kong
Unlike conventional contrast-weighted imaging, quantitative MR parametric maps can be obtained from MRF and should conceivably be very useful for the quantification and delineation of normal and pathologic tissues. Together with the fact that MRF is very efficient , our central hypothesis is that MRF is an ideal alternative to existing MRI motion tracking methods. In this study, we have demonstrated that it is possible to track motion by continuously performing MRF during free breathing. MR parametric maps for each respiratory phases were retrospectively estimated from the MRF snapshots that fall into a given respiratory bin.
|
|
3739.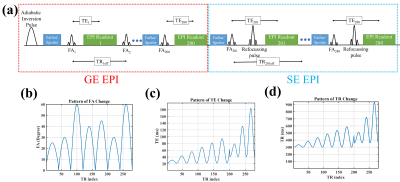 |
Slice Accelerated EPI-based Magnetic Resonance Fingerprinting for Simultaneous Estimation of T1, T2, T2* with Whole-brain Coverage
Mahdi Khajehim1,2, Thomas Christen3, Fred Tam4, Simon Graham1,4, and J. Jean Chen1,2
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Rotman Research Institute, Baycrest Health Sciences, Toronto, ON, Canada, 3Grenoble Institute of Neurosciences, Inserm, Grenoble, France, 4Hurvitz Brain Sciences Research Program, Sunnybrook Research Institute, Toronto, ON, Canada Magnetic resonance fingerprinting (MRF) offers a way to quantitatively estimate multiple relaxation parameters with a single scan, but fast MRF estimation of T1, T2, and T2* with a single acquisition remains challenging. Here, a dual-stage EPI-based MRF approach with online image reconstruction is proposed for estimating T1, T2, and T2*. We achieved 1.1x1.1x3 mm3 resolution with minimal distortion, and with the use of simultaneous multi-slice acceleration, our method can provide whole-brain coverage (1.7x1.7x3 mm3) in less than 3 minutes. |
|
3740.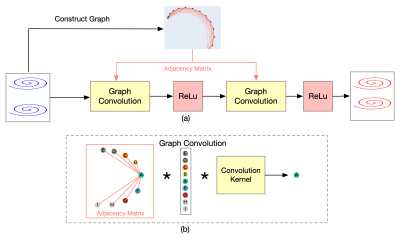 |
Accelerated High-Resolution 3D MR Fingerprinting Using a Graph Convolutional Network
Feng Cheng1, Zhenghan Fang2, Xiaopeng Zong3, Weili Lin3, Yong Chen3, and Pew-Thian Yap3
1Department of Computer Science, University of North Carolina at Chapel Hill, CHAPEL Hill, NC, United States, 2CuraCloud Corporation, Seattle, WA, United States, 3Department of Radiology, University of North Carolina at Chapel Hill, CHAPEL Hill, NC, United States
In this study, a k-space interpolation technique for high-resolution 3D MR Fingerprinting is proposed. We formulate the problem as a graph and apply a graph convolutional network on the graph to interpolate the missing partitions. Our preliminary results show that the proposed method can provide improved results both in reconstructed k-space data and in extracted quantitative maps and can potentially allow higher acceleration factors along the partition-encoding direction.
|
|
3741.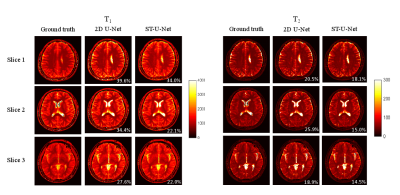 |
ST-UNet: Spatio-Temporal U-Net for Accelerating Simultaneous MultiSlice Magnetic Resonance Fingerprinting
Yilin Liu1, Xiaopeng Zong1, Zhenghan Fang1, Weili Lin1, Dinggang Shen1, Pew-Thian Yap1, and Yong Chen1
1University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
In this study, a spatio-temporal U-Net was developed for efficient and accurate T1 and T2 quantifications from multislice MR Fingerprinting acquisitions. Our preliminary results demonstrate that the proposed method outperforms the standard template matching method and deep learning methods in the literature, enabling higher multislice factors for MR Fingerprinting.
|
|
3742.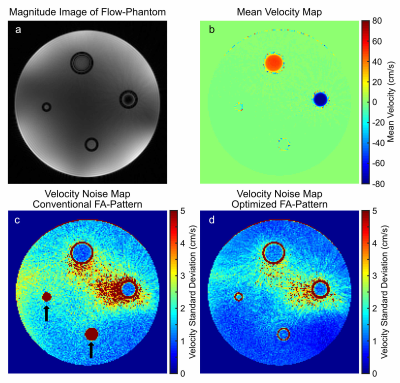 |
Flip Angle Optimization for Continuous Flow-MRF Acquisition under Periodic Boundary Conditions
Sebastian Flassbeck1, Fabian Kratzer1, Simon Schmidt1, Lisa Leroi2, Mark E. Ladd1, and Sebastian Schmitter1,2
1German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Physikalisch-Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany
Flow-MRF is a highly promising technique for rapid quantification of both relaxation times and time-resolved flow velocities. However, this method currently suffers from two major drawbacks, firstly a low efficiency since multiple shots are used and secondly strongly increased velocity noise for low velocities due to magnetization preparation pulses. This abstract targets both issues by presenting an optimized the flip angle (FA)-pattern for Flow-MRF, while assuming periodic boundary conditions.
|
|
3743.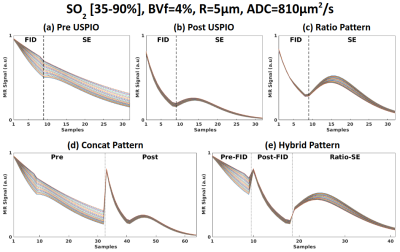 |
Optimizing signal patterns for MR vascular fingerprinting
Aurelien Delphin1, Fabien Boux1,2, Clément Brossard1, Jan M Warnking1, Benjamin Lemasson1, Emmanuel Luc Barbier1, and Thomas Christen1
1Univ. Grenoble Alpes, Inserm, U1216, Grenoble Institut Neurosciences, GIN, 38000, Grenoble, France, 2Univ. Grenoble Alpes, Inria, CNRS, G-INP, 38000, Grenoble, France
MR vascular fingerprinting proposes to map vascular properties such as blood volume fraction, average vessel radius or blood oxygenation saturation (SO2). The fingerprint pattern used in previous studies provides low sensitivity on SO2. We optimised signal patterns built from pre and post USPIO acquisitions. Concatenation of different echoes associated with higher dimensional dictionaries led to better estimates in both healthy and tumoral tissues.
|
|
3744.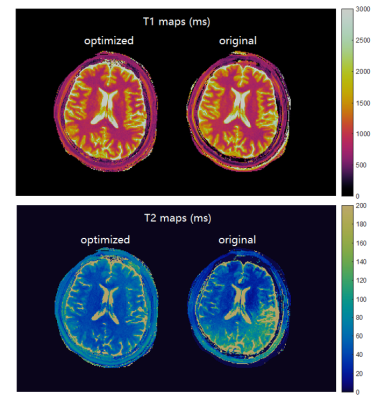 |
Quantum Optimization Framework for MR Fingerprinting Framework Incorporating Undersampling and Noise
Siyuan Hu1, Ignacio Rozada2, Rasim Boyacioglu3, Stephen Jordan4, Sherry Huang1, Matthias Troyer4, Mark Griswold3, Debra McGivney1, and Dan Ma1
1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 21Qbit, Vancouver, BC, Canada, 3Radiology, Case Western Reserve University, Cleveland, OH, United States, 4Microsoft, Redmond, WA, United States
MR fingerprinting is a novel quantitative MR imaging technique that provides multiple tissue properties maps simultaneously. Designing appropriate MR fingerprinting sequence patterns is crucial to speed up data acquisition while obtaining accurate measurements. Here we propose an advanced MR fingerprinting optimization framework that incorporates undersampling artifacts and random noise in the cost function which directly compute quantitative errors in the result maps. We use quantum-inspired algorithm to solve the problem and generate optimized sequences. In both simulation and in vivo experiments, the optimized sequence showed improved image quality and measurement accuracy.
|
|
3745.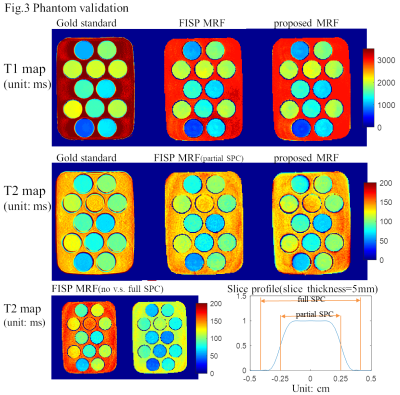 |
A generalized combined FISP and PSIF MR Fingerprinting with improved T2 quantification
Huihui Ye1,2, Qiqi Tong2, Qing Li2,3, Xiaozhi Cao2, Hongjian He2, Jianhui Zhong2, and Huafeng Liu1
1State Key Laboratory of Modern Optical Instrumentation, College of Optical Science and Engineering, Zhejiang University, Hangzhou, China, 2Center for Brain Imaging Science and Technology, Key Laboratory for Biomedical Engineering of Ministry of Education, College of Biomedical Engineering and Instrumental Science, Zhejiang University, Hangzhou, China, 3MR Collaborations, Siemens Healthcare Ltd., Shanghai, China
A generalized combined FISP and PSIF MR Fingerprinting sequence is proposed where either FISP block or PSIF block can be used in each TR. An example sequence pattern shows improved T2 mapping accuracy and its immunity to slice profile imperfection.
|
|
3746.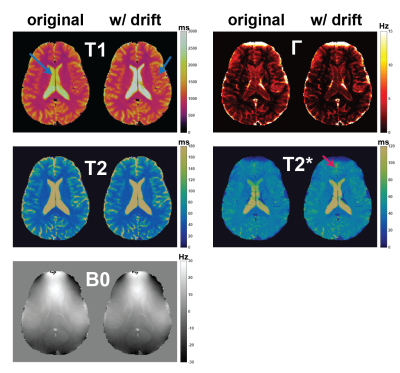 |
Magnetic Resonance Fingerprinting Reconstruction and Dictionary Matching with Compensation of Frequency Drifts
Rasim Boyacioglu1 and Mark Griswold1
1Radiology, Case Western Reserve University, Cleveland, OH, United States
Magnetic Resonance Fingerprinting (MRF) maps multiple tissue properties and system parameters simultaneously. The accuracy of MRF maps depends on the simulation of all possible system properties into the signal evolutions via the Bloch equations. We have observed frequency drifts during MRF scans, similar to those seen in fMRI scans, which might cause various artifacts if not accounted for. Here, it is shown that 2D MRF frequency drifts can be compensated with a simple dictionary update. For correction of 3D MRF frequency drifts, a novel reconstruction framework is introduced. Results show significant improvements on quantitative maps for 2D and 3D MRF.
|
|
3747.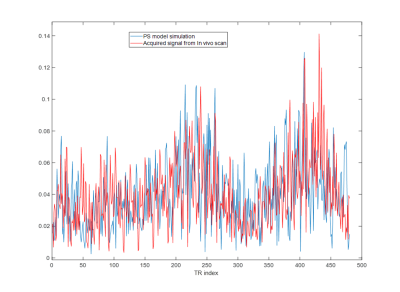 |
A Fast Approximation of Undersampling Artifacts in MR Fingerprinting
Debra McGivney1, Rasim Boyacıoğlu2, Stephen Jordan3, Ignacio Rozado4, Sherry Huang1, Siyuan Hu1, Brad Lackey3, Matthias Troyer3, Mark Griswold2, and Dan Ma1
1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Radiology, Case Western Reserve University, Cleveland, OH, United States, 3Microsoft, Redmond, WA, United States, 41QB Information Technologies, Vancouver, BC, Canada Poster Permission Withheld
Iterative optimization in MRI is a large problem with many degrees of freedom. Depending on the cost function and parameters of interest, it may be beneficial to model errors from undersampling with non-Cartesian trajectories. This typically requires repeated use of the nonuniform FFT (NUFFT), which is computationally expensive. Here we propose an approximation based on a limited number of tissue types that eliminates the need for repeated NUFFTs, and allows a wide range of applications for sequence optimization in MRI and MRF.
|
|
3748.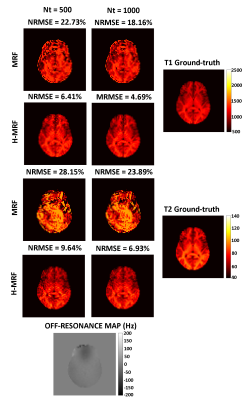 |
Reducing off-resonance effects by spatial decorrelation in Magnetic Resonance Fingerprinting
Ronal Manuel Coronado1,2,3, Gastão Lima da Cruz4, Claudia Prieto1,2,4, and Pablo Irarrázaval1,2,3,5
1Biomedical Imaging Center, Pontificia Universidad Catolica de Chile, Santiago, Chile, 2Department of Electrical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 3Millennium Nucleus for Cardiovascular Magnetic Resonance, Chile, Santiago, Chile, 4School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, London, United Kingdom, 5Institute for Biological and Medical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, Santiago, Chile
Magnetic Resonance Fingerprinting (MRF) acquisitions based on balanced Steady State Free Precession (bSSFP) with spiral trajectories produces higher signal noise to ratio (SNR) in comparison to the unbalanced version, nevertheless its usage is hider because of higher sensitivity to off-resonance artifacts. These artifacts affect the quality of parametric values. Here we propose a novel method to decrease these susceptibility distortions by means of spatial decorrelation. We show promising results where off-resonance artifacts were reduced in a realistic brain simulation and standardized T1/T2 phantom acquisition.
|
|
3749.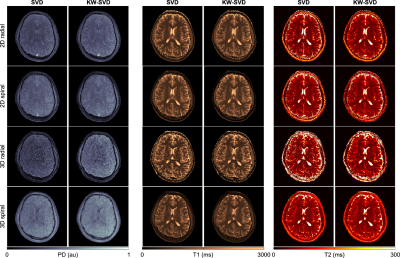 |
Effects of spatial encoding strategies on 2D and 3D magnetic resonance fingerprinting
Matteo Cencini1,2, Pedro A Gómez3, Mohammad Golbabaee 4, Rolf F Schulte5, Giada Fallo1,2, Luca Peretti1,2, Michela Tosetti2,6, Bjoern H Menze3, and Guido Buonincontri2,6
1University of Pisa, Pisa, Italy, 2Imago7 Foundation, Pisa, Italy, 3Technical University of Munich, Munich, Germany, 4University of Bath, Bath, United Kingdom, 5GE Healthcare, Munich, Germany, 6IRCCS Stella Maris, Pisa, Italy
Transient-state imaging techniques such as MR Fingerprinting allow for simultaneous quantification of tissue properties by using variable acquisition parameters in conjunction with undersampled non-Cartesian trajectories. Several implementations exist in literature, relying on two- or three-dimensional sampling readouts. Here, we studied the effect of the spatial encoding on quantification. In addition, an evaluation of the impact on parametric maps of anti-aliasing techniques (k-space weighted image constrast) was performed, both in vitro and in vivo.
|
|
3750.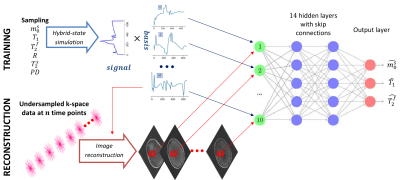 |
Optimized dimensionality reduction for parameter estimation in MR fingerprinting via deep learning
Quentin Duchemin1, Kangning Liu1, Carlos Fernandez-Granda2, and Jakob Assländer3
1Center for Data Science, NYU, New York, NY, United States, 2Courant Institute of Mathematical Sciences and Center for Data Science, New York University, New York, NY, United States, 3Center for Biomedical Imaging, Department of Radiology, New York University School of Medicine, Center for Advanced Imaging Innovation and Research (CAI2R), Department of Radiology, New York University School of Medicine, New York, New York, NY, United States
We propose a deep learning approach for MR fingerprinting that jointly learns a low-dimensional representation of the fingerprints and estimates biophysical parameters from this subspace. In contrast to SVD-based projections, which are agnostic to the estimation task, the learned subspace is optimized to maximize information content about the parameters of interest. Incorporating the learned basis functions in the forward imaging operator suppresses undersampling artifacts and increases computational efficiency.
|
|
3751.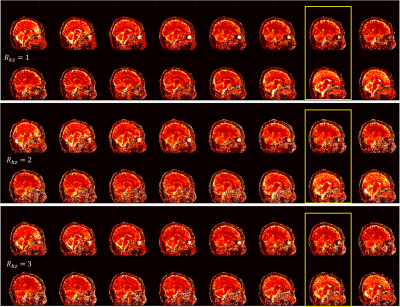 |
Accelerated 3D Magnetic Resonance Fingerprinting Using Alternating Direction Method of Multipliers
Di Cui1, Xiaoxi Liu1, Peng Cao1, Queenie Chan2, and Edward S. Hui1
1Diagnostic Radiology, The University of Hong Kong, Hong Kong, China, 2Philips Healthcare, Hong Kong, Hong Kong
3D magnetic resonance fingerprinting was developed for volumetric parametric quantification. In this study, an alternating direction method of multipliers (ADMM) based 3D MRF approach is proposed to jointly utilize sparsity constraint and spatial coil sensitivity information, leading to better reconstruction performance with 3-fold undersampled 3D MRF data. We demonstrated that the effective scan time for R=3 and whole-brain MRF is less than 7 minutes.
|
3752.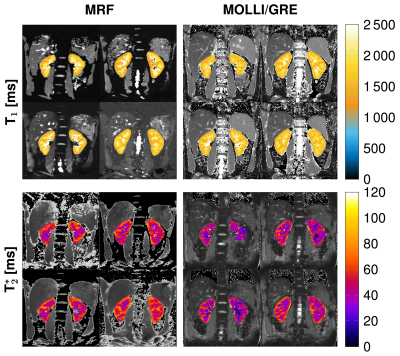 |
Renal magnetic resonance fingerprinting for simultaneous T1 and T2* quantification
Ingo Hermann1,2, Jorge Chacon-Caldera1, Irène Brumer1, Sebastian Weingärtner2, Lothar R. Schad1, and Frank G. Zöllner1
1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany, 2Magnetic Resonance Systems Lab, Department of Imaging Physics, Delft University of Technology, Delft, Netherlands
We implemented and validated a MRF sequence for simultaneous T1 and T2* quantification in the kidneys covering 4 slices within one breath-hold. Therefore, we used an echo-planar-imaging readout with fully sampled k-space and 35 measuremets for the matching process. We achieved promising results in phantom and in 8 healthy volunteers compared with conventional methods as MOLLI and GRE and with reference scans. Additionally, we performed smoothing on the baseline images to further improve the image qualitiy of the MRF parametric maps.
|
|
3753.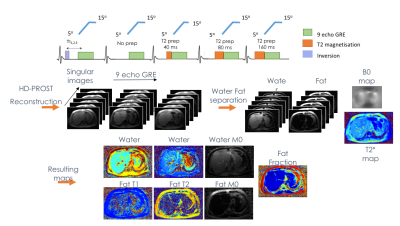 |
Validation of a water and fat separation framework for Liver MR Fingerprinting
Cristobal Arrieta1,2, Olivier Jaubert3, Gastao Cruz3, Sergio Uribe1,2,4, Rene M Botnar3, Claudia Prieto3, and Carlos Sing-Long2,5,6
1Biomedical Imaging Center, Pontificia Universidad Catolica de Chile, Santiago, Chile, 2Millennium Nucleus for Cardiovascular Magnetic Resonance, Santiago, Chile, 3School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 4Radiology Department, School of Medicine, Pontificia Universidad Catolica de Chile, Santiago, Chile, 5Instituto de Ingeniería Matemática y Computacional, Pontificia Universidad Catolica de Chile, Santiago, Chile, 6Millennium Nucleus Center for the Discovery of Structures in Complex Data, Santiago, Chile
In this work we validated a liver MR Fingerprinting technique which allows to simultaneously estimate T1, T2, Proton Density Fat Fraction (PDFF) and T2*. This technique is based on a novel water and fat separation optimisation scheme, which includes l2-norm of the fieldmap gradient, TV regularisation on T2* and l1-norm denoising of the water and fat concentrations. We presented results on T1/T2 and water/fat phantoms and in 11 volunteers. This approach showed to be robust and it is ready to be applied on more clinical challenging cases.
|
|
3754. |
Simultaneous Multi-contrast Four-dimensional Magnetic Resonance Imaging for Radiotherapy Applications
Tian Li1, Di Cui2, Edward S. Hui2, and Jing Cai1
1The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 2The University of Hong Kong, Hong Kong, Hong Kong
Tumor motion imaging is of vital importance in managing mobile cancers in radiation therapy. However, current 4D-MRI techniques are inefficient and ineffective, potentially leading to suboptimal and inaccurate results. To solve this problem, we propose a novel four-dimensional magnetic resonance fingerprinting (4D-MRF) technique for radiation therapy applications. Our proposed method has been validated through simulations and in-vivo volunteer experiment.
|
|
| 3755. | The effect of hormone therapy on T1 mapping-related values obtained from MR fingerprinting-derived images of prostate cancer patients
Nikita Sushentsev1, Joshua D Kaggie1, Guido Buonincontri2,3, Rolf Schulte4, Vincent J Gnanapragasam5,6,7, Martin J Graves1, and Tristan Barrett1,8
1Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 2IRCCS Stella Maris, Piza, Italy, 3Imago7 Foundation, Piza, Italy, 4GE Healthcare, Munich, Germany, 56) Cambridge Urology Translational Research and Clinical Trials Office, University of Cambridge, Cambridge, United Kingdom, 6Academic Urology Group, Department of Surgery & Oncology, University of Cambridge, Cambridge, United Kingdom, 7Department of Urology, University of Cambridge, Cambridge, United Kingdom, 82) CamPARI Prostate Cancer Group, Addenbrooke's Hospital, Cambridge, United Kingdom
This study investigates the effect of hormone therapy for prostate cancer on T1 mapping values obtained from MR Fingerprinting. No differences were observed between tumour T1 values before and after treatment. However, lower T1 MRF values were noted in prostate lesions compared to normal peripheral and transition zones, which is consistent with current literature. Moreover, a significant decrease in T1 values was observed in normal transition zone (TZ) following treatment, which may justify the need of using T1 mapping in contrast-enhanced MRI studies aimed at calculating quantitative TZ-derived parameters.
|
|
3756. |
Quantitative Tracking of T1 and T2* in DCE-MRI by Dynamic MR Fingerprinting
Xueying Zhao1, Ying-Hua Chu2, Qianfeng Wang1, and He Wang1,3
1Institute of Science and Technology for Brain-inspired Intelligence, Fudan University, Shanghai, China, 2MR Collaboration, Siemens Healthcare Ltd., Shanghai, China, 3Human Phenome Institute, Fudan University, Shanghai, China
Precise quantification of the contrast agent uptake in DCE-MRI is still challenging. Here, we redesigned the reconstruction scheme of MR fingerprinting by introducing a sliding window into the process of dictionary matching, which allows dynamic quantification of T1 and T2* in DCE-MRI with high temporal resolution. The performance of this proposed dynamic MRF method is simulated and tested on MATLAB. The time-varying trajectories of T1 and T2* have been successfully captured with a temporal resolution of 4.5 seconds. This method may open the door of utilizing MR fingerprinting to quantify time-varying variable with adjustable temporal resolution.
|
|
3757.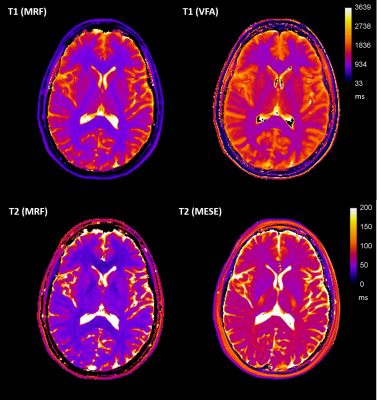 |
Validity and reproducibility of Magnetic Resonance Fingerprinting in the healthy human brain at 3T
Joely Smith1,2, Ben Statton3, Sarah Cardona1, Mary Elizabeth Finnegan1,2, Rebecca Abigail Quest1,2, and Matthew Grech-Sollars1,4
1Department of Imaging, Imperial College Healthcare NHS Trust, London, United Kingdom, 2Department of Bioengineering, Imperial College London, London, United Kingdom, 3MRC London Institute of Medical Sciences, Imperial College London, London, United Kingdom, 4Department of Surgery and Cancer, Imperial College London, London, United Kingdom
Obtaining quantitative measurements in a 7-minute acquisition could improve the sensitivity of MR diagnosis. We investigated the validity and reproducibility of magnetic resonance fingerprinting (MRF) relaxometry in 12 tissue compartments in the human brain through comparison to standard mapping techniques: variable flip angle for T1 and multi-echo spin echo for T2. Statistically significant strong and moderate correlations were found between the MRF and standard mapping methods for T1 and T2, respectively. The MRF results were shown to be highly reproducible and in agreement with values found within the literature. However, a bias was found between MRF and standard relaxometry methods.
|
|
3758.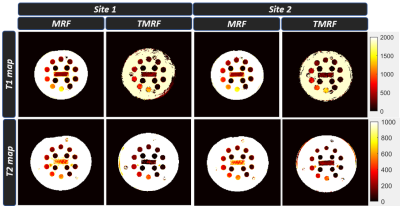 |
Two-site repeatability study of Tailored Magnetic Resonance Fingerprinting (TMRF)
Enlin Qian1, Amaresh Shridhar Konar2, Pavan Poojar1, Maggie Fung3, and Sairam Geethanath1
1Columbia Magnetic Resonance Research Center, New York, NY, United States, 2Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3GE Healthcare Applied Sciences Laboratory East, New York, NY, United States
This work evaluates repeatability of T1 and T2 estimation of the TMRF method compared to MRF and the gold standard measurements at two sites. In this work, we acquired data for 10 days on ISMRM/NIST phantom using the three methods at the two sites with the same vendor and field strength. The data was reconstructed and matched with an EPG simulated dictionary. ROI analysis was performed to extract T1 and T2 estimation for each sphere. The results and statistical analysis are presented here.
|
|
3759.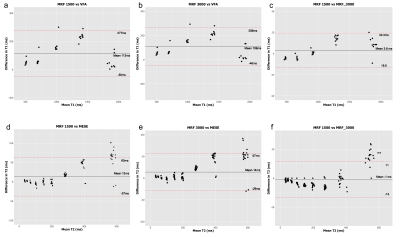 |
Accuracy, reproducibility and temperature variability of Magnetic Resonance Fingerprinting using the ISMRM/NIST system phantom
Ben Statton1,2, Joely Smith3,4, Mary E Finnegan3,4, Rebecca A Quest3,4, and Matthew Grech-Sollars3,5
1Imperial College London, London, United Kingdom, 2Medical Research Council, London Institute of Medical Sciences, London, United Kingdom, 3Department of Imaging, Imperial College Healthcare NHS Trust, London, United Kingdom, 4Department of Bioengineering, Imperial College London, London, United Kingdom, 5Department of Surgery and Cancer, Imperial College London, London, United Kingdom
Magnetic Resonance Fingerprinting (MRF) is a technique which produces multiple parametric maps during a single fast acquisition. Before MRF can be adopted clinically, quantitative values derived from these maps must be proven accurate and reproducible over a range of T1 and T2 values and temperatures. The aim of this study was to investigate the accuracy and reproducibility of T1and T2 values derived from two different methods of MRF compared to conventional quantitative maps using the ISMRM/NIST system phantom.
|
|
3760.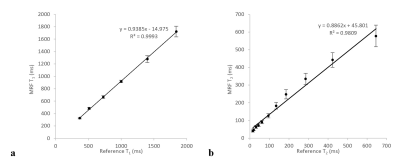 |
Repeatability of Magnetic Resonance Fingerprinting using ISMRM/NIST MRI Phantom in Philips 3T MRI Scanner
Varut Vardhanabhuti1, Ho Ting Au2, Jie Ding1, Elaine Y Lee1, Peng Cao1, and Edward S Hui1
1Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong, 2The University of Hong Kong, Hong Kong, Hong Kong
The purpose of this study is to assess the repeatability of the magnetic resonance fingerprinting (MRF) sequence in a Philips 3T scanner with reference to the ISMRM/NIST MRI system phantom. The results showed a strong correlation between the T1 and T2 estimated from MRF versus the reference values (R2 = 0.999, and R2 = 0.981). A high level of repeatability was achieved over the 15 scanning sessions, although the T1 estimated from MRF has a significantly higher degree of repeatability than T2.
|
|
3761.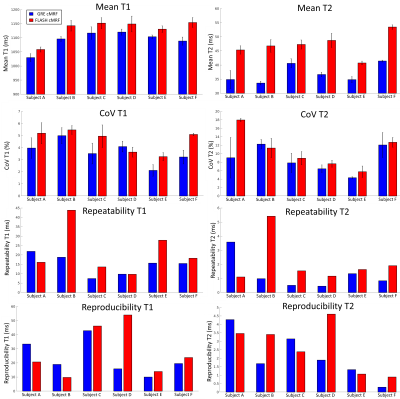 |
Comparing FLASH vs GRE for 2D cardiac MR fingerprinting
Gastao Cruz1, Olivier Jaubert1, Aurelien Bustin1, Torben Schneider2, Peter Koken3, Mariya Doneva3, René M. Botnar1, and Claudia Prieto1
1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Philips Healthcare, Guildford, United Kingdom, 3Philips Research Hamburg, Hamburg, Germany
2D Cardiac Magnetic Resonance Fingerprinting (cMRF) has been previously proposed to provide co-registered T1/T2 maps from a single breath-hold scan. Gradient Spoiled gradient echo (GRE) readout is conventionally employed for 2D cMRF. RF-spoiled readouts (FLASH) have reduced signal-to-noise ratio but are less sensitive to field inhomogeneities that may bias the parametric maps (if not encoded in the MRF sequence). Here we compare the performance of FLASH and GRE readouts for simultaneous T1/T2 mapping in 2D cMRF. Phantom and in-vivo results showed that both sequences produce comparable maps and reproducible values, however GRE-cMRF presented larger underestimations for T1 and T2.
|
|
3762.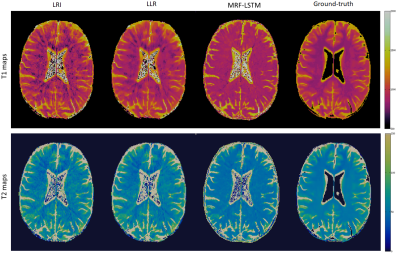 |
Tissue based denoising for MR fingerprinting via long short-term memory networks
Gastao Cruz1, Thomas Kuestner1, Ilkay Oksuz1, Olivier Jaubert1, Niccolo Fuin1, Andy P. King1, Julia A. Schnabel1, René M. Botnar1, and Claudia Prieto1
1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Conventional Magnetic Resonance Fingerprinting (MRF) relies on pixelwise dictionary matching of highly undersampled time-series images. However, remaining aliasing artefacts in these images can compromise the matching step and thus affect the accuracy of the parametric maps. Dictionary-based compression has been proposed to exploit redundancies in the signal evolution dimension, however these approaches do not exploit tissue redundancies within the images. Here we propose to leverage redundant information between similar tissues and the MRF dictionary to suppress residual artefacts along time, using long short-term memory (LSTM) networks. Preliminary results indicate that proposed MRF-LSTMs can suppress aliasing in highly undersampled scenarios.
|
|
3763.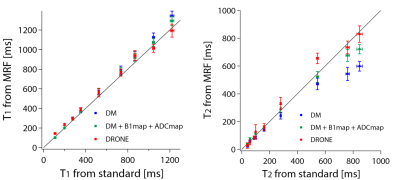 |
Accurate estimation of multiple parameters from MRF signals using deep learning
Ryoichi Sasaki1 and Yasuhiko Terada1
1Institute of Applied Physics, University of Tsukuba, Tsukuba, Japan Ideally, MRF can quantify multiple parameters at a single scan. In some cases, however, these parameters are not separable and additional scans for inseparable parameters are required, which reduces the advantage of the short scan time of MRF. This is remarkable when the large number of parameters are involved in the signal evolution process. Here, we used a pattern matching using a deep neural network called DRONE, and verified the separability of four parameters of T1, T2, B1, and ADC from MRF-FISP signals acquired at a single scan. |
|
3764.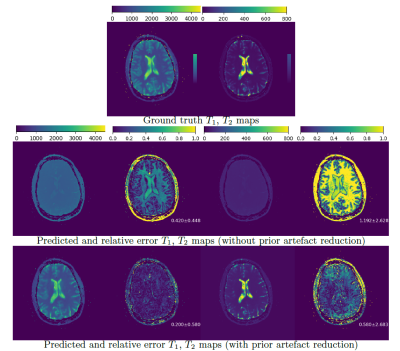 |
Learning how to Clean Fingerprints -- Deep Learning based Separated Artefact Reduction and Regression for MR Fingerprinting
Yiling Xu1, Elisabeth Hoppe1, Peter Speier2, Thomas Kluge2, Mathias Nittka2, Gregor Körzdörfer2, and Andreas Maier1
1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare, Erlangen, Germany
Various deep learning approaches have been recently introduced to enable a fast MRF reconstruction compared to dictionary matching. Artefacts resulting from the strong undersampling during the acquisition often impair the reconstruction results. In this work, we introduce a deep learning artefact reduction method in order to provide clean fingerprints for the subsequent regression network. Our results achieve a decreased relative error by over 50% using our artefact reduction method compared to previously proposed deep learning regression model without prior artefact reduction.
|
|
3765.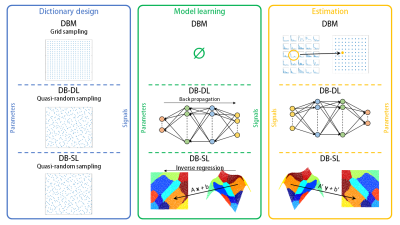 |
Dictionary-based Learning in MR Fingerprinting: Statistical Learning versus Deep Learning
Fabien Boux1,2, Florence Forbes2, Julyan Arbel2, Aurélien Delphin1, Thomas Christen1, and Emmanuel L. Barbier1
1Univ. Grenoble Alpes, Inserm, U1216, Grenoble Institut Neurosciences, GIN, 38000, Grenoble, France, 2Univ. Grenoble Alpes, Inria, CNRS, G-INP, 38000, Grenoble, France
In MR Fingerprinting, the exhaustive search in the dictionary may be bypassed by learning a mapping between fingerprints and parameter spaces. In general, the relationship between these spaces is particularly non-linear, which implies the use of advanced regression methods: deep learning frameworks but also methods based on statistical models have been proposed. In this study, we compare reconstruction time, accuracy and noise robustness of the conventional dictionary-matching method and two methods that handle the modelling of the non-linear relashionship with a neural network and a statistical inverse regression model.
|
|
3766.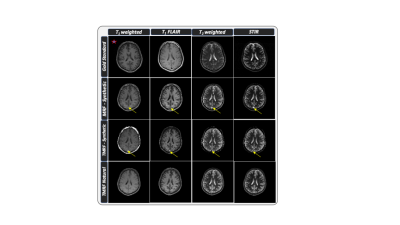 |
Natural, multi-contrast and quantitative imaging of the brain using tailored MR fingerprinting
Pavan Poojar1,2, Enlin Qian1, Maggie Fung3, and Sairam Geethanath1,2
1Columbia University Magnetic Resonance Research Center, Columbia University in the City of New York, New York, NY, United States, 2Dayananda Sagar College of Engineering, Bangalore, India, 3GE Healthcare Applied Sciences Laboratory East, New York, New York, NY, United States
Tailored MRF (TMRF) provides scheme to acquire non-synthetic, multi contrast images simultaneously in one sequence by tailoring the acquisition scheme derived in MRF. Natural contrast obtained from TMRF overcomes artifacts observed in MRF such as flow and errors in tissue parameter quantitation. We acquired TMRF and MRF data on the ISMRM/NIST phantom along with gold standard relaxometry data. We also acquired MRF and TMRF data for 5 volunteers and compared the natural and synthetic contrasts; and the relaxometric maps. The TMRF natural contrasts are similar to control experiments and do not contain flow artifacts found in synthetic contrasts.
|
|
3767.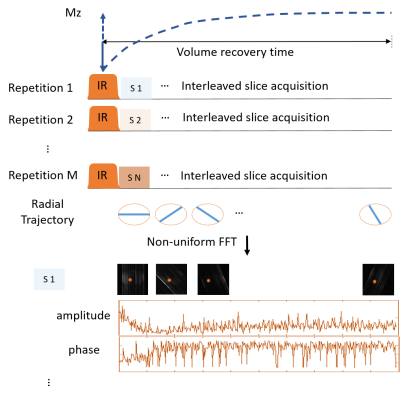 |
Fast T1 mapping with inversion recovery followed by radial acquisition and dictionary-matching reconstruction
Qing Li1, Xiaoyue Zhou1, and Yi Sun1
1MR Collaborations, Siemens Healthcare Ltd., Shanghai, China A dictionary-based reconstruction method was applied to estimate T1 mapping from a series of highly undersampled images acquired with an inversion recovery–prepared FISP sequence and a radial readout. Motion robustness improved using interleaved slice acquisition and data rejection in T1 estimation. The measurement time was 4 s/slice. Phantom and in vivo knee results show an up to 50% data rejection results in less than 10% of quantification accuracy loss, compared to the T1 map reconstructed with the full dataset. |
 |
3768.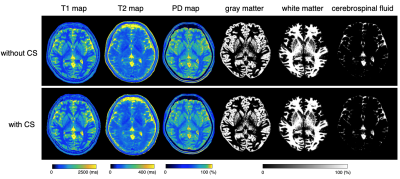 |
Compressed sensing applied to 1 mm isotropic multi-parametric imaging with 3D-QALAS: A phantom, volunteer, and patient study
Shohei Fujita1,2, Akifumi Hagiwara1, Naoyuki Takei3, Ken-Pin Hwang4, Issei Fukunaga1, Shimpei Kato1,2, Masaaki Hori5, Ryusuke Irie1,2, Christina Andica1, Toshiaki Akashi1, Koji Kamagata1, Ukihide Tateishi6, Osamu Abe2, and
Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3MR Applications and Workflow, GE Healthcare, Tokyo, Japan, 4Department of Radiology, MD Anderson Cancer Center, Houston, TX, United States, 5Department of Radiology, Toho University Omori Medical Center, Tokyo, Japan, 6Department of Radiology, Tokyo Medical and Dental University, Tokyo, Japan
Simultaneous relaxometry of T1, T2, and proton density by 3D-QALAS still requires relatively lengthy acquisition times and further acceleration is desired in clinical practice. Here, we applied compressed sensing combined with parallel imaging to 3D-QALAS to accelerate scan time. Quantitative values and tissue segmentation performance were compared with and without acceleration in phantom, healthy volunteers, and patients. With accelerated 3D-QALAS acquisition, whole brain 1 mm isotropic volume data with multiple parametric maps and co-registered tissue segmentation maps were obtained in less than 6 minutes, with comparable quality to 3D-QALAS without acceleration.
|
3769.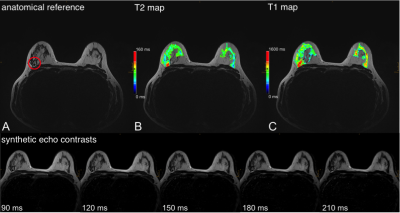 |
Quantitative and Synthetic MRI for Breast Assessment in Clinical Acquisition Times
Nina Pötsch1, Marcus Raudner1,2, Tom Hilbert3,4,5, Tobias Kober3,4,5, Elisabeth Weiland6, Panagiotis Kapetas1, and Pascal Baltzer1
1Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 3Advanced Clinical Imaging Technology, Siemens Healthcare, Lausanne, Switzerland, 4Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 5LTS5, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 6Siemens Healthcare GmbH, Erlangen, Germany
Contrast-enhanced breast MRI is the most sensitive tool for the detection of breast cancer. Contrast enhancement is however not specific to cancer: incidental benign findings regularly require further workup including ultimately avoidable follow-up scans and biopsies. Quantitative T1 and T2 measurements could be used as discriminative imaging markers to assist clinical decision-making. This feasibility study illustrates the potential clinical benefits of quantitative imaging for breast MRI by combining two fast and robust mapping techniques for high-resolution parameter mapping in a clinically feasible scan time of 7:32 min using prototype compressed sensing MP2RAGE and GRAPPATINI sequences with a harmonized protocol.
|
|
3770.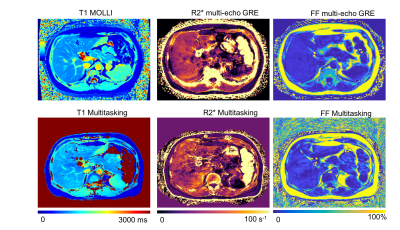 |
Three-Dimensional Free-breathing Whole-Liver Simultaneous T1, R2*, and Fat-Fraction Quantification Using MR Multitasking
Nan Wang1,2, Anthony G Christodoulou1, Yibin Xie1, Fei Han3, Xiaodong Zhong3, Sen Ma1,2, Xiaoming Bi3, Vibhas Deshpande4, and Debiao Li1
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 3Siemens Healthcare, Los Angeles, CA, United States, 4Siemens Healthcare, Austin, TX, United States
Quantitative MRI has been playing a key role in the assessment and characterization of liver. However, current applications are still facing technical challenges. In this work, we proposed a novel technique based on MR Multitasking, which enables simultaneous T1, R2*, and FF quantification in one scan with free-breathing acquisition, 3D whole-liver coverage, and sufficient spatial resolution. The feasibility of the proposed method were demonstrated on the volunteer study (N = 8), showing that the T1, R2*, and FF quantification was repeatable in vivo and consistent with the results from the reference methods.
|
|
3771.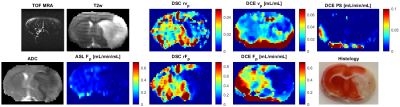 |
Blood Flow and Permeability Imaging in a Rat Stroke Model Using 3D Compressed-Sensing DCE-MRI
Radovan Jiřík1, Lucie Krátká1, Jiří Kratochvíla1, Ondřej Macíček1, Ye Tian2, Peter Scheer3, Jana Hložková3, Edward DiBella2, and Zenon Starčuk, jr.1
1Institute of Scientific Instruments of the CAS, Brno, Czech Republic, 2Utah Center for Advanced Imaging and Research, University of Utah, Salt Lake City, UT, United States, 3University of Veterinary and Pharmaceutical Sciences Brno, Brno, Czech Republic
The main challenge of using DCE-MRI in stroke in combination with advanced pharmacokinetic models is the need for high temporal resolution, whole-brain coverage and a low SNR (especially in small-animal MRI) of the DCE signal (compared to DSC, due to the BBB and low fractional blood volume in brain). In this pilot study we test if compressed sensing could help solve these challenges.
|
|
3772.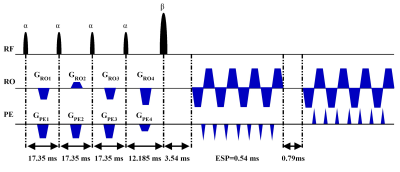 |
Snapshot Quantitative MRI: Application to Simultaneously T2 and T2* Mapping
Jian Wu1, Xiaoyin Wang2, Hongjian He2, Lingceng Ma1, Shuhui Cai1, Congbo Cai1, and Jianhui Zhong3
1Department of Electronic Science, Xiamen University, Xiamen, China, 2The Center for Brain Imaging Science and Technology, Zhejiang University, Zhejiang, China, 3Department of Imaging Sciences, University of Rochester, Rochester, NY, United States
Simultaneously quantifying T2 and T2* properties can provide great sensitivity and specificity to diseases. However, most of the existing methods are time-consuming. Here, we develop an overlapping-echo method for simultaneously achieving reliable T2 and T2* maps in a single shot. This method is robust to motion and the inhomogeneity of B0. Experimental results demonstrate that the resulting T2 and T2* values are in good agreement with those obtained with reference methods.
|
|
3773.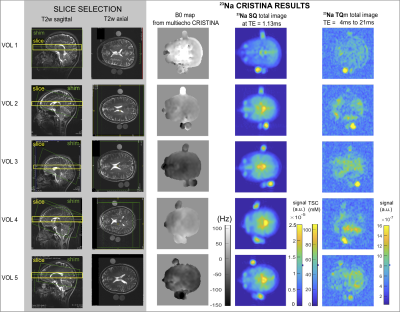 |
CRISTINA: Cartesian Single and Triple Quantum Imaging of 23Na in the Brain
Michaela A U Hoesl1, Lothar R Schad1, and Stanislas Rapacchi2
1Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 2Aix-Marseille University, CRMBM, CNRS, Marseille, France
Multi-quantum sodium imaging offers additional insights compared to standard single-quantum (SQ) images with information beyond tissue sodium concentration (TSC). Simulation including B0 inhomogeneity and stimulated echo signals of multi-quantum signals and spectra of phase cycle choices facilitated robust sequence development. The characteristic temporal evolution of the SQ and triple quantum (TQ) sodium is of interest and a multi echo sequence for SQ and TQ signal acquisition was developed, which was evaluated in phantom and 5 healthy volunteers in the brain.
|
|
3774.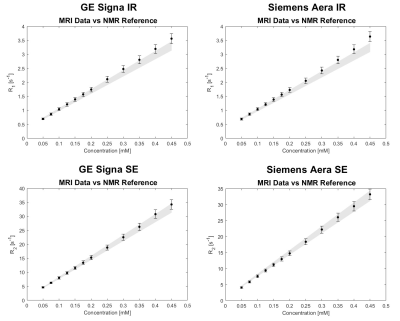 |
Phantom study on magnetic resonance imaging (MRI) T1 and T2 relaxation times measurements standardization
Davide Cicolari1, Domenico Lizio2, Patrizia Pedrotti3, Monica Teresa Moioli2, Alessandro Lascialfari1, Manuel Mariani1, and Alberto Torresin2,4
1Department of Physics, Università degli Studi di Pavia, Pavia, Italy, 2Department of Medical Physics, ASST Grande Ospedale Metropolitano Niguarda, Milan, Italy, 3Department of Cardiology, ASST Grande Ospedale Metropolitano Niguarda, Milan, Italy, 4Department of Physics, Università degli Studi di Milano, Milan, Italy
Relaxation times measurement standardization, a great issue in clinical inter-centre and inter-scanner applications, is studied by comparing relaxation times maps of a MnCl2 phantom, scanned with two different MRI imagers, with reference values measured with an NMR laboratory spectrometer. For this study standard sequences were used: NMR reference T1 and T2 values were obtained from IR (Inversion-Recovery) and CPMG (Carr-Purcell-Meiboom-Gill) sequences respectively; MRI maps were generated from clinical IR and SE (Spin-Echo) sequences. The MRI and NMR results agreement within the experimental error limits (5%) suggests that the estimation of the relaxation times is independent from the spectrometer/scanner utilized.
|
|
3775.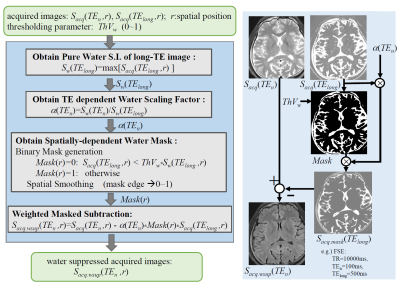 |
Synthetic MRI with T2-based Water Suppression without Loss of Tissue SNR
Tokunori Kimura1, Kousuke Yamashita1, and Kouta Fukatsu1
1Department of Radiological Science, Shizuoka College of Medicare Science, Hamamatsu-Shi, Japan
We proposed a modified T2-based water suppression synthetic-MRI technique without loss of tissue SNR, which was introduced by subtraction of heavy-T2W image from standard acquired images. Our water suppression was achieved by subtracting only water portions except for tissue portions. We demonstrated both effects that CSF-PVE artifacts were dramatically suppressed and the tissue SNR was kept to before subtraction in our water suppressed quantitative maps; and thus water suppressed synthetic images of FLAIR and SE provided better gray-white matter contrasts than those for subtracting uniformly.
|
|
3776.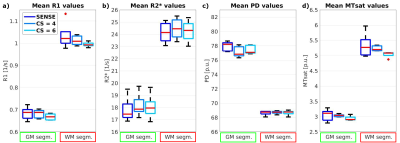 |
Evaluating Compressed SENSE acceleration for multi-parametric quantitative mapping of R1, R2*, PD, and MTsat with the hMRI toolbox
Ronja Berg1, Tobias Leutritz2, Stephan Kaczmarz1, Claus Zimmer1, Nikolaus Weiskopf2, and Christine Preibisch1
1School of Medicine, Department of Neuroradiology, Technical University of Munich, Munich, Germany, 2Max Planck Institute for Human Cognitive and Brain Sciences, Department of Neurophysics, Leipzig, Germany
Measuring physical parameters quantitatively by magnetic resonance imaging (MRI) has a high value for diagnostic applications as it allows the detection of disease related systemic changes. However, quantitative MRI mapping methods increase the scan time significantly compared to conventional imaging. Therefore, we investigated the applicability of Compressed SENSE (CS) acceleration in a multi-parametric mapping protocol for R1, R2*, PD, and MTsat imaging. Our results demonstrate that absolute parameter values remained constant in all evaluated regions-of-interest when applying CS. Thus, CS can be used to almost halve the scan for R1, R2*, PD, and MTsat mapping without loss of fidelity.
|
|
3777.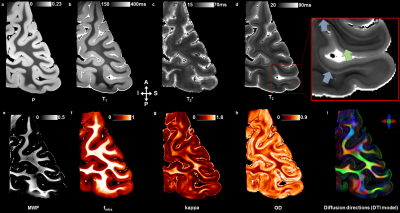 |
Ex vivo mapping of the cyto- and the myeloarchitecture of the human cerebral cortex using ultra-high field MRI (7T and 11.7T)
Raïssa Yebga Hot1,2, Alexandros Popov1,2, Justine Beaujoin1, Gaël Perez1,3, Fabrice Poupon1,2, Igor Lima Maldonado4, Jean-François Mangin1,2, Christophe Destrieux4, and Cyril Poupon1,2
1CEA - NeuroSpin, Gif-sur-Yvette, France, 2Université Paris-Saclay, Orsay, France, 3CentraleSupélec, Gif-sur-Yvette, France, 4Imaging and Brain laboratory (iBrain), Université de Tours - INSERM, Tours, France
The investigation of the human cerebral cortex at the mesoscopic scale remains challenging but promising to better understand brain pathologies associated with cortex damage. In this ex vivo study, samples of occipital cortex from both hemispheres of an unique subject have been delineated using their microstructural and myeloarchitectural information inferred from ultra-high field anatomical, quantitative and diffusion MRI. The high-resolved UHF-MRI dataset enabled to perform an automatic segmentation of the cortical layers within the primary and secondary visual cortices. The segmentations highlighted their commonalities and differences between the two hemispheres.
|
|
3778.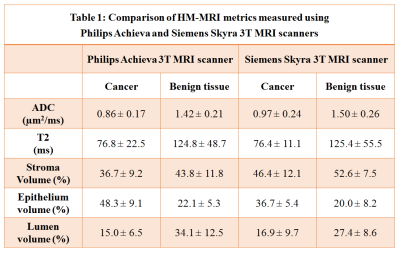 |
Cross vendor validation of Hybrid Multidimensional MRI in the non-invasive measurement of prostate tissue composition
Aritrick Chatterjee1, Grace Lee2, Deb Dietz2, Aytekin Oto1, and Gregory Karczmar1
1Department of Radiology, University of Chicago, Chicago, IL, United States, 2Ingalls Memorial Hospital, Flossmoor, IL, United States
This study evaluates the consistency of HM-MRI for non-invasive measurement of prostate tissue composition on scanners from two MRI vendors. HM-MRI was performed on Philips Achieva 3T (with endorectal coil) and Siemens Skyra 3T (without endorectal coil) MRI scanners. HM-MRI metrics measured for cancerous and benign prostatic tissue using Philips and Siemens were similar, with slight variation due to different patients in each cohort. Diagnostic accuracy for detecting PCa using HM-MRI was similar for both MR vendors: Philips (AUC = 0.94-0.99, p<0.05) and Siemens (AUC = 0.83-0.98, p<0.05).
|
|
3779.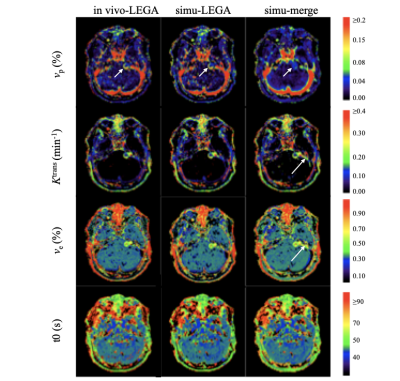 |
Accuracy assessment of high-spatial resolution whole-brain tracer-kinetic parameter maps generated using dual-temporal resolution DCE-MRI
Ka-Loh Li1, Daniel Lewis2, Sha Zhao1, Alan Jackson1, and Xiaoping Zhu1
1Division of Informatics, Imaging and Data Sciences, The University of Manchester, Manchester, United Kingdom, 2Manchester Centre for Clinical Neurosciences, Salford Royal NHS Foundation Trust, Manchester, United Kingdom
Accurate, high-spatial resolution whole-brain pharmacokinetic maps are highly desirable in clinical neuro-oncology practice. A new dual-temporal resolution based kinetic mapping technique for this purpose, termed LEGATOS, was recently described and tested with in-vivo patient data. In this study, quantitative assessment of the accuracy of parameter estimates derived using the LEGATOS analysis procedure was evaluated through computer simulation. Structural similarity and percentage deviation of the “measured” values from the known “true” values were used for evaluation and demonstrated that the LEGATOS technique offered superior accuracy compared to the use of unreconstructed composite HT and HS curves alone.
|
|
3780.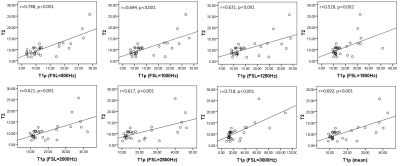 |
T1ρ could be a complementary tool for T2* in the assessment of rat liver iron overload
Qianfeng Wang1, Hong Xiao2, He Wang1, Xuchen Yu1, and Fuhua Yan2
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China In this study, spin-lock and 2D UTE pulse sequences were developed to quantify T1ρ and T2* of 32 rat livers with iron overload at 11.7T MR system. Moreover, T2 was also acquired to compare with T1ρ and T2*. Pearson correlation analyses (two-tailed) illustrated that T1ρ were significantly associated with T2 (all p ≤ 0.002), but insignificantly related with T2* (all p > 0.05). No significant association was found between T2 and T2*. |
|
3781. |
Simultaneous B1- and Fat-Corrected T1 Mapping Using Chemical-Shift Encoded MRI
Nathan Tibbitts Roberts1,2, Diego Hernando1,2,3,4, Timothy Colgan1, Daiki Tamada1, and Scott B Reeder1,3,4,5,6
1Radiology, University of Wisconsin - Madison, Madison, WI, United States, 2Electrical and Computer Engineering, University of Wisconsin - Madison, Madison, WI, United States, 3Medical Physics, University of Wisconsin - Madison, Madison, WI, United States, 4Biomedical Engineering, University of Wisconsin - Madison, Madison, WI, United States, 5Medicine, University of Wisconsin - Madison, Madison, WI, United States, 6Emergency Medicine, University of Wisconsin - Madison, Madison, WI, United States
Spatially varying B1 inhomogeneities and tissue fat are known confounders of quantitative T1 mapping methods that use variable flip angle techniques. Separately acquired B1 calibration maps can be used to correct flip angle errors caused by B1 inhomogeneities, but this requires an additional acquisition. In this work we propose a novel approach for simultaneous estimation of B1, T1, proton density fat-fraction and R2* using dual orthogonal RF pulses and multiple flip angles. The feasibility and noise performance of this proposed acquisition and fitting strategy are evaluated using Cramer-Rao Lower Bound analysis, simulations, and preliminary phantom experiments.
|
|
3782.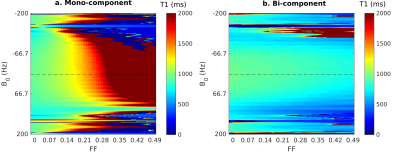 |
Increasing robustness to fat partial volume effects in T1-mapping with MOLLI
Andreia S Gaspar 1, Andreia C Freitas1, and Rita G Nunes1
1ISR-Lisboa/LARSyS and Department of Bioengineering, Instituto Superior Técnico – Universidade de Lisboa, Lisbon, Portugal
Modified Look-Locker inversion recovery (MOLLI) with a balance Steady State Free Precession (bSSFP) readout is widely applied in the clinical setting but it is known to be sensitive to fat partial volume effects (PVE), as well as static B0 field inhomogeneities (ΔB0). As an alternative, the FLASH readout, more robust to ΔB0, can be applied for T1 mapping. In this work we studied the robustness of FLASH-MOLLI to fat PVE, and propose a new bi-component model to improve the accuracy of water T1 estimates in both bSSFP and FLASH MOLLI.
|
|
3783.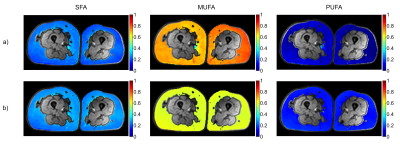 |
In Vivo Validation of MRS- and MRI-Based Fatty Acid Composition Quantification against Gas Chromatography in Adipose Tissue
Lena Trinh1, Pernilla Peterson1,2, Håkan Brorson3, and Sven Månsson1,4
1Medical Radiation Physics, Department of Translational Medicine, Lund University, Malmö, Sweden, 2Medical Imaging and Physiology, Skåne University Hospital, Lund, Sweden, 3Department of Plastic and Reconstructive Surgery, Skåne University Hospital, Malmö, Sweden, 4Radiation Physics, Skåne University Hospital, Malmö, Sweden Non-invasive estimation of the fatty acid composition of adipose tissue using MRI or MRS may be valuable in a number of disease scenarios. However, in vivo validation against an independent gold standard is still needed. In this work, we find that especially MRI estimation of the fraction of saturated fatty acids is strongly associated to gas chromatography analysis in the adipose tissue of lymphedema patients. The reliability of the estimated mono- and polyunsaturated fractions are reliant on the used signal model. Also MRS provided results which are associated to GC analysis, but with a lower agreement compared to MRI. |
3784.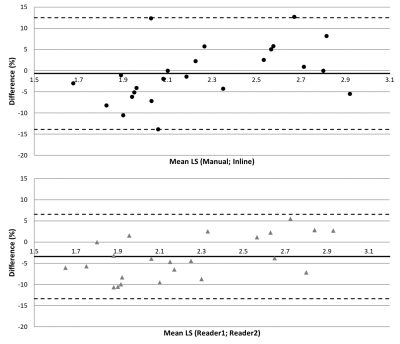 |
Initial Experience with Inline Evaluation of Liver Stiffness using Magnetic Resonance Elastography in Hemochromatosis Patients
Stephan A.R. Kannengiesser1, Cathy Cazin2, Michel Lapp2, Khalid Ambarki3, Berthold Kiefer1, and Valérie Laurent2,4
1MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 2Department of Radiology, CHRU Nancy, Brabois Adults Hospital, Vandoeuvre-lès-Nancy, France, 3Siemens Healthcare SAS, Saint Denis, France, 4IADI, U1254, INSERM, Université de Lorraine, CHRU de Nancy Brabois, Vandoeuvre-lès-Nancy, France A simple prototype inline mean stiffness evaluation approach for MR Elastography based on anatomical liver segmentation and the confidence map was evaluated in 25 hemochromatosis patients, and compared with manual evaluation by two readers according to QIBA guidelines. Pairwise comparisons were found to be of similar statistics: mean±std relative difference -3.40±5.10% and -0.69±6.72% for reader1 vs. reader2 and manual (average of readers) vs. inline. The presented approach is promising given the particular patient cohort and limited stiffness range, but further evaluation is needed. |
|
3785.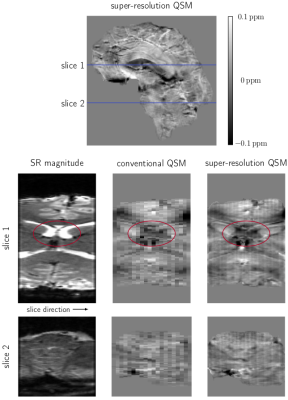 |
Fast, high resolution QSM of the brainstem at 7T using super-resolution 2D EPI
Andreas Ehrmann1, Beata Bachrata1,2, Korbinian Eckstein1, David Bancelin1, Pedro Lima Cardoso1, Charles Joseph Guthrie1, Siegfried Trattnig1,2, and Simon Daniel Robinson1,3,4
1High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Christian Doppler Laboratory for Clinical Molecular MR Imaging, Vienna, Austria, 3Department of Neurology, Medical University of Graz, Graz, Austria, 4Centre for Advanced Imaging, University of Queensland, Queensland, Australia
2D EPI-based Quantitative Susceptibility Mapping offers a means to localise brainstem structures very quickly. However, this comes at the price of limited achievable slice thickness, restricting the precision of the localisation. In this work, we develop a fast super-resolution reconstruction method for complex MRI data, allowing the reconstruction of high resolution magnitude and phase images, and hence also high resolution susceptibility maps. We demonstrate that the reconstructed super-resolution 2D EPI images significantly improve the visibility of small structures allowing for improved localisation of the brainstem and other small structures such as veins in the generated susceptibility maps.
|
|
3786.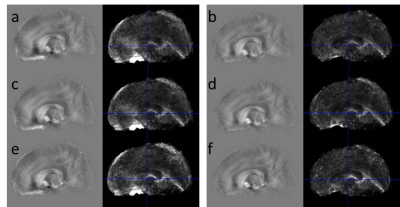 |
Towards robust QSM in cortical and sub-cortical regions of the human brain at 9.4T: influence of coil combination and masking strategies
Gisela E Hagberg1,2, Korbinian Eckstein3, Enrique Cuna4, Simon Robinson3,5, and Klaus Scheffler1,2
1Biomedical Magnetic Resonance, University Hospital Tübingen, Tübingen, Germany, 2High Field Magnetic Resonance, Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 3High Field MR Centre, Medical University of Vienna, Vienna, Austria, 4Centro Uruguayo de Imagenología Molecular (CUDIM), Montevideo, Uruguay, 5Centre for Advanced Imaging, University of Queensland, Brisbane, Australia
Strong background signals leading to multiple phase wraps may hamper accurate quantification of magnetic tissue susceptibility (QSM) especially at high field strengths using long echo times to achieve a high spatial sampling. Here we show how different coil-combination, automated tissue masking and background removal techniques can be used to improve QSM quality. Performance was evaluated with regard to iron quantification in subcortical and cortical areas in the same subjects. We found a substantial improvement in accuracy and precision of QSM in high-field applications at long echo times through the use of ASPIRE and removal of areas with excessive phase evolution.
|
|
3787.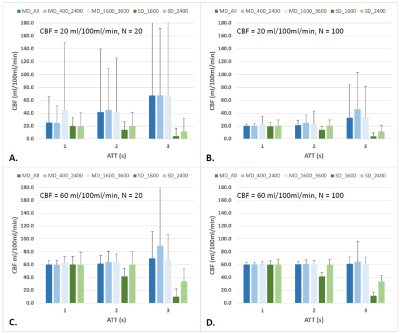 |
The cerebral blood flow derived from multi-delay ASL MRI varies with the choice of post-labeling delay
Ya-Fang Chen1, Sung-Chun Tang2, and Wen-Chau Wu3,4
1Department of Medical Imaging, National Taiwan University Hospital Hsin-Chu Branch, Hsinchu, Taiwan, 2Department of Neurology, National Taiwan University Hospital, Taipei, Taiwan, 3Institute of Medical Device and Imaging, National Taiwan University, Taipei, Taiwan, 4Graduate Institute of Clinical Medicine, National Taiwan University, Taipei, Taiwan
With arterial spin labeling (ASL) MRI, the delay time between labeling and image acquisition is a critical imaging parameter for accurate flow quantification. Multi-delay ASL has been proposed to tackle the problem of unknown arterial transit time (ATT). However, it is not fully understood how the range of delay times affects the derived CBF. The present study aimed to investigate this potential issue. Results show that the CBF derived from multi-PLD ASL varies with the choice of PLD (overestimated when the PLD range does not cover ATT) as well as SNR (overestimated along with greater variability when SNR is low).
|
|
3788.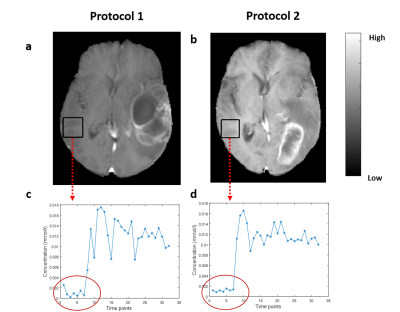 |
To Evaluate the Effect of Compressed-SENSE on Accuracy of High-Resolution Quantitative T1-Perfusion MRI Parameters and on glioma grading at 3T
Dinil Sasi S1, Rakesh K Gupta2, Indrajit Saha3, Anandh K Ramaniharan3, and Anup Singh4,5
1Indian Institute of Technology Delhi, New Delhi, India, 2Fortis Memorial Research Institute, Gurugram, India, 3Philips India Limited, Gurugram, India, 4Indian Institute of Technology Delhi, Hauz Khas, India, 5All India Institute of Medical Science, New Delhi, India
T1-perfusion derived parameters have been utilized for brain tumor quantification and grading. In this study, we have analyzed the potential of Compressed SENSE (CSENSE) acceleration technique on high resolution T1-perfusion MRI for quantitative analysis of brain tumor(glioma) and compared its performance and accuracy with conventional acquisition protocol in terms of concentration-time curve, perfusion parameters and glioma grading. A prospective analysis was also carried on healthy subjects to analyze the spatial error propagation at different acceleration factors(R). All derived perfusion parameters were able to quantify and differentiate different tumor classes with improved concentration-time curve(High grade and low grade glioma).
|
|
3789.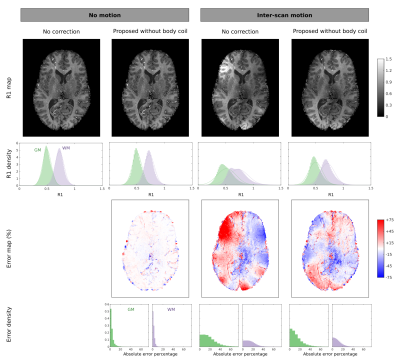 |
Estimation of net receive sensitivity - at 3T and 7T - for correction of inter-scan motion artefacts in R1 mapping
Yaël Balbastre1, Nadège Corbin1, and Martina F Callaghan1
1Wellcome Centre for Human Neuroimaging, University College London, London, United Kingdom
The longitudinal relaxation rate is a useful myelin proxy that can be estimated by combining multiple acquisitions with different nominal flip angles. Inter-scan motion can introduce large biases that can be efficiently corrected if reference data with a relatively ‘flat’ sensitivity profile, e.g. a body coil, is available. This is generally not the case at ultra-high field where highly inhomogeneous localised transmit-receive coils are used. Therefore, we propose a new method for estimating and removing the net coil sensitivity that does not require a body coil, needs only coil-wise magnitude images, yet reduces motion-induced bias in R1.
|
|
3790.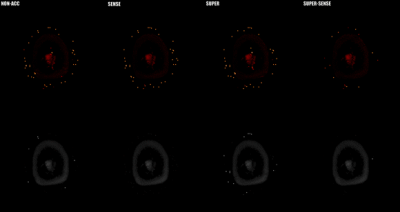 |
Acceleration of 3D high-resolution variable-flip-angle T1 mapping based on SUPER
Fan Yang1, Jun Xie2, Guobin Li2, Meng Jiang3, and Chenxi Hu4
1College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China, 2United Imaging Healthcare Co., Ltd, Shanghai, China, 3Department of Cardiology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China, 4Institute of Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Three-dimensional variable-flip-angle(VFA) T1-mapping is a rapid T1 quantification method with important clinical applications. However, the requirement of acquiring multiple 3D images greatly increases the scan time. We propose a novel application of SUPER—a recently developed framework for parametric mapping acceleration—to reduce scan time and/or improve spatial resolution of 3D VFA T1-mapping. In healthy subjects, we demonstrate that SUPER(R=2) and SUPER-SENSE(R=4 combining SUPER and parallel imaging) achieved similar accuracy, reasonable noise amplification, and similar reconstruction time compared with the non-acceleration gold standard. The whole upper-brain T1-mapping scan time was reduced from 5.33 minutes to 1.33 by employment of SUPER-SENSE.
|
|
3791.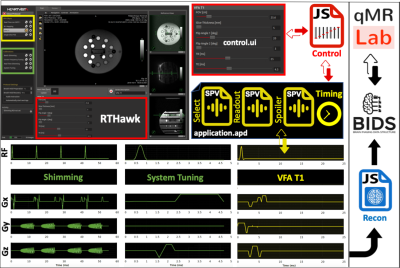 |
Thinking outside the black box: A fully transparent T1 mapping pipeline
Agah Karakuzu1,2, Mathieu Boudreau1,2, Julien Cohen-Adad1,3, and Nikola Stikov1,2
1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Heart Institute, Montreal, QC, Canada, 3Unité de Neuroimagerie Fonctionnelle (UNF), Centre de recherche de l’Institut Universitaire de Gériatrie de Montréal (CRIUGM), Montreal, QC, Canada
Quantitative MRI is pointless if we cannot peak inside the black box that generates the numbers. This is the main motivation behind our concept for a fully transparent qMRI pipelines starting at the scanner console and extending all the way to a journal publication. In this work, we developed a simple application to demonstrate this for variable flip angle (VFA) T1 mapping. This application exemplifies the feasibility of a vendor-neutral qMRI sequence that is publicly shared under version control (https://github.com/qMRLab/pulse_sequences). Our approach opens a way towards community-driven development and improvement of qMRI applications without the need for specialized programming skills.
|
|
3792.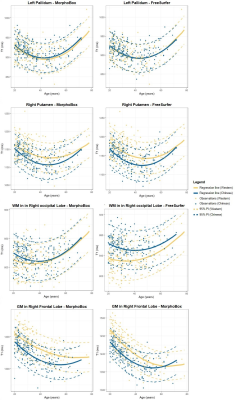 |
Brain T1 relaxometry changes across the life span: a comparison between two populations
Gian Franco Piredda1,2,3, Peipeng Liang4, Tom Hilbert1,2,3, Karl Egger5, Shan Yang5, Jean-Philippe Thiran2,3, Bénédicte Maréchal1,2,3, Yi Sun6, Kuncheng Li7,8, and Tobias Kober1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4School of Psychology, Capital Normal University, Beijing Key Laboratory of Learning and Cognition, Beijing, China, 5Faculty of Medicine, University of Freiburg, Freiburg, Germany, 6MR Collaboration, Siemens Healthcare Ltd., Shanghai, China, 7Department of Radiology, Xuanwu Hospital, Capital Medical University, Beijing, China, 8Beijing Key Laboratory of Magnetic Resonance Imaging and Brain Informatics, Beijing, China
Understanding the normal evolution of T1 values across the life span allows to disentangle ageing from degenerative pathologies. Following previous studies reporting brain anatomical and functional differences between Western and Chinese cohorts, this work investigates whether differences in the evolution of T1 values across the life span exist between these two populations using two datasets with 200 healthy subjects each. Derived trends were found to differ between the two populations in some brain structures, especially in grey matter tissues. The observed differences may indicate that norms derived from one population may not be directly applied to another without recalibration.
|
|
3793.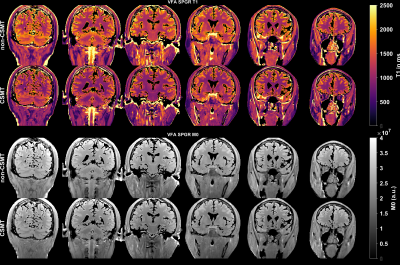 |
Variable flip angle T1 mapping with MT-balanced RF pulses
Nam Gyun Lee1, Zhibo Zhu2, and Krishna S. Nayak2
1Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States
Quantitative DCE requires accurate and precise pre-contrast M0/T1 maps. Variable flip angle (VFA) T1 mapping has good precision but has accuracy issues. Magnetization transfer (MT) effects is one of the major reasons for differences between VFA and reference IR-FSE T1 measurements. Here, we apply the recent framework for MT-balanced RF pulse design to high-resolution whole-brain T1 mapping.
|
|
3794.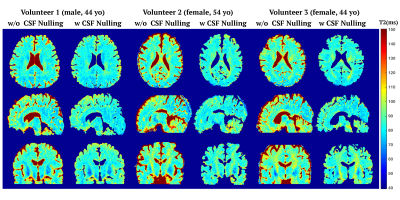 |
Quantitative T2 Mapping using Accelerated 3D Stack-of-Spiral GRE Acquisition
Ruoxun Zi1, Dan Zhu1,2, Wenbo Li2,3, and Qin Qin2,3
1Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
We presented a T2 prepared stack-of-spiral gradient echo (GRE) pulse sequence with or without CSF nulling for T2 mapping of brain. T2 maps were iteratively optimized by data consistency, model consistency and spatial sparsity regularization using projected-gradients approach, which was evaluated by in vivo experiments and compared with SENSE, CS SENSE and model-based methods. The proposed reconstruction method provided better performance with normalized root mean square error (nRMSE) of the whole brain T2 estimation equal to 8.2±0.5% with an acceleration factor of 5. The T2 estimation with and without CSF nulling were consistent.
|
|
3795. |
Joint reconstruction and estimation for fast and accurate T2* mapping
Seonyeong Shin1, Riwaj Byanju2, Seong Dae Yun1, Stefan Klein2, Dirk H. J. Poot2, and N. Jon Shah1,3,4,5
1Institute of Neuroscience and Medicine 4, INM-4, Forschungszentrum Jülich, Jülich, Germany, 2Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 3Institute of Neuroscience and Medicine 11, INM-11, JARA, Forschungszentrum Jülich, Jülich, Germany, 4JARA - BRAIN - Translational Medicine, Aachen, Germany, 5Department of Neurology, RWTH Aachen University, Aachen, Germany
Quantification of T2* is relatively time-efficient. However, still the scan times might be too long for the desired resolution. In this work, we demonstrated the ability to accelerate the acquisition by joint reconstruction and parameter estimation for T2* quantification. From retrospectively highly subsampled k-spaces, images and T2* maps were reconstructed directly. It was shown that an acceleration factor of 16 is feasible for T2* mapping.
|
|
3796.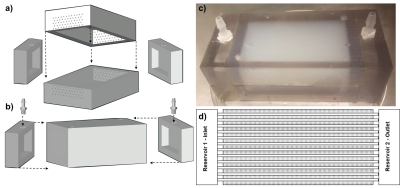 |
A 3D-Printed Multi-level Perfusion Phantom for Quantitative Analysis of Perfusion Models vs. Experimental Data
John Morgan1 and Yi Wang1
Video Permission Withheld
1Cornell University, New York, NY, United States
Current methods for perfusion characterization are difficult to quantify absolutely and provide only relative and qualitative information. Perfusion-phantoms that enable quantitative analysis of transport phenomena are needed to test theoretical models against experimental data. The initial design and deployment of a 3D-printed phantom with pump-driven perfusion of multi-level microvascular structure encapsulated in hydrogel is presented. It simulates vascularized tissue and enables experimental validation. A new quantitative analytical method , using voxelized constitutive convection-diffusion equations, is applied to DCE data and compared to traditional Kety’s methods. The largely qualitative and unmeasurable Kety global AIF assumption is replaced with measurable and reproducible data.
|
|
3797.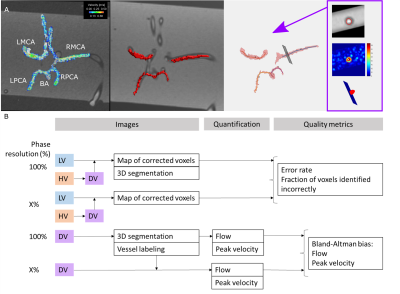 |
Variable Spatial Resolution Dual-Venc 4D Flow MRI: Balancing Image Quality and Scan Time
Maria Aristova1, Jianing Pang2, Liliana Ma1, Michael Markl1, and Susanne Schnell1
1Radiology, Northwestern University, Chicago, IL, United States, 2Siemens Healthcare, Chicago, IL, United States
Dual-venc 4D flow MRI has previously been reported to achieve wide velocity dynamic range while maintaining a high velocity-to-noise ratio, which is particularly relevant for neurovascular applications. To increase flexibility in spatiotemporal resolution, we present a prototype implementation of a interleaved 8-point dual-venc 4D Flow acquisition with independently prescribed (prospectively undersampled) spatial resolution of the high venc acquisition, i.e. Variable Spatial Resolution Dual Venc (VSRDV). This allowed anti-aliasing error rates less than 15% error for a high venc spatial resolution of 70-80%. This represents 10-15% reduction of scan time.
|
|
3798.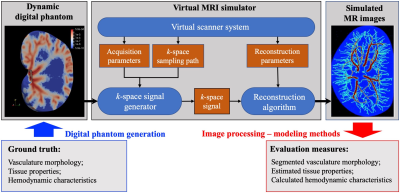 |
Validation of dynamic contrast-enhanced MRI analyses via virtual MRI simulation on a dynamic digital phantom
Chengyue Wu1, Ty Easley2, Victor Eijkhout3, David A. Hormuth4, Federico Pineda5, Gregory S. Karczmar5, and Thomas Yankeelov1,4,6,7
1Department of Biomedical Engineering, University of Texas at Austin, Austin, TX, United States, 2Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO, United States, 3HPC Software Tools Group, Texas Advanced Computing Center, Austin, TX, United States, 4Oden Institute for Computational Engineering and Sciences, University of Texas at Austin, Austin, TX, United States, 5Department of Radiology, University of Chicago, Chicago, IL, United States, 6Department of Diagnostic Medicine, University of Texas at Austin, Austin, TX, United States, 7Department of Oncology, University of Texas at Austin, Austin, TX, United States
We have recently developed novel image processing and computational methods to characterize both the morphology and hemodynamics of tumor-associated vessels to assist in the diagnosis of suspicious breast lesions. In this contribution, we employ a dynamic digital phantom of contrast agent perfusion and extravasation to systematically evaluate the sensitivity of our methods to spatial resolution, temporal resolution, and signal-to-noise ratio (SNR). The immediate goal is to use the phantom to determine an acquisition-reconstruction protocol that can implemented in the routine clinical setting, thereby enabling quantitative hemodynamic analysis to be widely available for the characterization of cancer.
|
|
3799.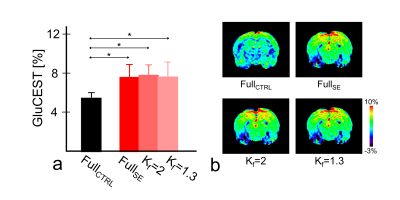 |
Optimization of Keyhole Imaging Parameters for Glutamate Chemical Exchange Saturation Transfer (GluCEST) MRI at 7.0 T
Dong-Hoon Lee1, Do-Wan Lee2, Chul-Woong Woo3, Hwon Heo4, Jae-Im Kwon3, Yeon Ji Chae4, Su Jung Ham5, Jeong Kon Kim2, Kyung Won Kim2,5, and Dong-Cheol Woo3,4
1Faculty of Health Sciences and Brain & Mind Centre, The University of Sydney, Sydney, Australia, 2Department of Radiology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea, 3Convergence Medicine Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Republic of Korea, 4Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea, 5Asan Image Research, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Republic of Korea
We evaluated the effects of a reference image and keyhole factor selections for high-frequency substitution on a keyhole imaging technique for application in glutamate CEST (GluCEST) imaging to reduce data acquisition time. The calculated GluCEST signals and visually inspected results from the reconstructed GluCEST maps indicated that a combination of unsaturated image as a reference image and >50% of keyhole factors showed consistent signals and image quality as opposed to the fully-sampled CEST data. Combining the keyhole imaging technique with GluCEST imaging enables stable image reconstruction and quantitative evaluation, and this approach is potentially implemented in various CEST imaging applications.
|
3800.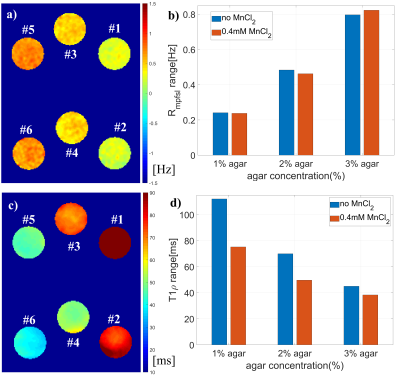 |
Quantitative Macromolecular Proton Fraction Imaging Based On Spin-Lock
Jian Hou1, Vincent Wong2, Baiyan Jiang1, Yixiang Wang1, Anthony Chan3, Winnie Chu1, and Weitian Chen1
1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 2Department of Medicine & Therapeutics, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 3Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong, Hong Kong, Hong Kong
Macromolecular Proton Fraction (MPF) is the relative amount of protons associated with macromolecules involved in magnetization transfer with free water protons. MPF is typically measured by quantitative magnetization transfer methods. In this work, we reported that MPF can also be measured based on spin-lock. Compared to the existing MPF methods, our method requires fewer parameter maps. We demonstrated our method using simulation, phantom, and in vivo experiments.
|
|
3801.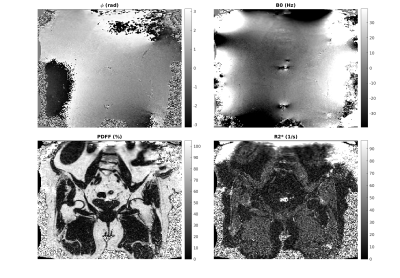 |
Constraints in Estimating the Proton Density Fat Fraction
Mark Bydder1, Vahid K Ghodrati1, Yu Gao1, Matthew D Robson2, Yingli Yang1, and Peng Hu1
1UCLA, Los Angeles, CA, United States, 2Perspectum Diagnostics, Oxford, United Kingdom
The study evaluates four physically motivated constraints in the estimation of the proton density fat fraction (PDFF) based on the physics of magnetic resonance imaging. These were smooth fieldmap, smooth initial phase, nonnegative proton density and moderate $$$R2*$$$ values. Results show that constraints are effective at reducing standard deviation and bias.
|
|
3802.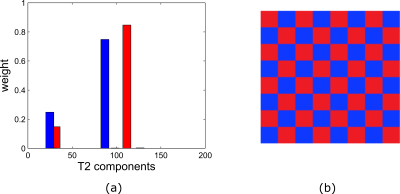 |
Faster Myelin water mapping by joint reconstruction and estimation from highly undersampled 3D-MSE acquisitions.
Riwaj Byanju1, Stefan Klein1, and Dirk H. J. Poot1
1Department of Radiology and Nuclear Medicine, Erasmus Medical Center, Rotterdam, Netherlands
Myelin water fractions (MWF) can provide biomarkers for many brain disorders. Multiple-spin-echo (MSE) is considered as gold standard for obtaining MWF. However, its clinical application is limited by its long acquisition time. Efficient undersampling and exploiting the redundancy in the contrast images of MSE can decrease the acquisition time. We propose a subspace based image reconstruction technique designed for reconstructing contrast images from highly undersampled MSE acquisition, so that it can be used for MWF mapping. We evaluate it for different signal-to-noise ratios for up to acceleration factor of 16 and show its feasibility for accelerating acquisition beyond parallel imaging.
|
|
3803.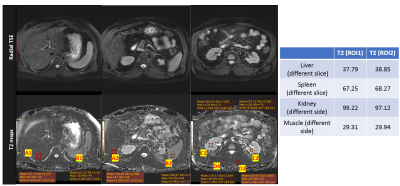 |
Improved T2 Quantification using Noise Corrected Stimulated Echo Compensation
Fei Han1, Mahesh Bharath Keerthivasan2, and Vibhas Deshpande3
1MR R&D and Collaboration, Siemens Healthineers, Los Angeles, CA, United States, 2MR R&D and Collaboration, Siemens Healthineers, Tucson, AZ, United States, 3MR R&D and Collaboration, Siemens Healthineers, Austin, TX, United States
Quantitative imaging is the key to developing reliable and reproducible imaging methods for standardized diagnostic exams. Most practical T2 mapping solutions are based on the CPMG concept, using the SEMC or TSE sequence. However, several confounding variables, such as, but not limited to the B1, RF profile, refocusing flip-angle, choice of TEs, and the imaging noise/artifacts, can lead to distorted signal and inaccurate T2 quantification. In this work, we investigated some of these confounders in CPMG-based T2 quantification and proposed a new model and fitting methods to improve the reliability, reproducibility of in-vivo T2 quantification.
|
|
3804.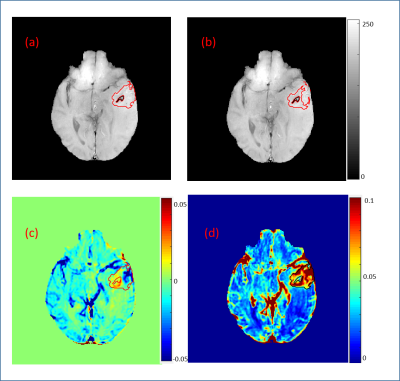 |
Method for Automatic blood vessel removal from quantitative T1-perfusion MRI maps and evaluating its impact on tumor grading
Manish Awasthi1, Bansmita Kar1, Neha Vats1, Virendra Kumar Yadav1, Dinil Sasi1, Mamta Gupta2, Rakesh Kumar Gupta2, and Anup Singh1,3
1Centre for Biomedical Engineering, Indian Institute of Technology, Delhi, New Delhi, India, 2Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 3Biomedical Engineering, All India Institute of Medical Science, Delhi, New Delhi, India
Presence of large blood vessels within tumor region can mislead interpretation on quantitative T1-perfusion MRI, particularly using automatic classification approaches. Purpose of this study was to develop a methodology for automatic blood vessel removal from quantitative T1-perfusion maps, compare it with previously reported methodology and finally evaluating impact of blood-vessel removal on tumor grading. In the proposed approach, signal intensity time curves characteristics, particularly contrast wash-out rate and peak value provided accurate automatic removal of blood-vessel from tumor region. Significant differences between T1-perfusion maps with and without blood-vessel removal were observed and tumor grading were also influenced.
|
|
3805.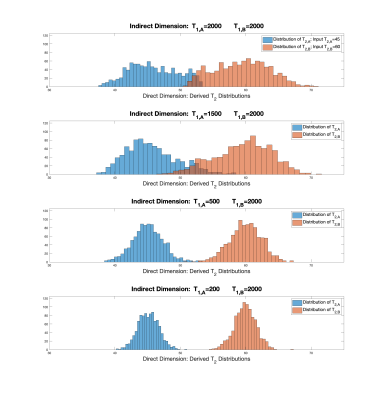 |
Statistical Analysis of Parameter Estimation for Multiexponential Decay in MR Relaxometry in One, Two, and Higher Dimensions
Richard G Spencer1, Mustapha Bouhrara1, Kenneth W Fishbein1, and Joy You Zhuo1
1National Institute on Aging, NIH, Baltimore, MD, United States
Analysis of multiexponential decay has remained a topic of active research for over 200 years, attesting to the difficulty in deriving the rate constants and component amplitudes of the underlying monoexponential decays. We have shown previously that parameter estimates from 2D relaxometry, with two distinct time variables, exhibit substantially greater accuracy and precision than those from analysis of 1D data, with a single time variable. We now present a statistical study of this remarkable phenomenon and indicate applications in 2D relaxometry and related experiments. These results provide a potential means of circumventing conventional limits on multiexponential parameter estimation.
|
|
3806.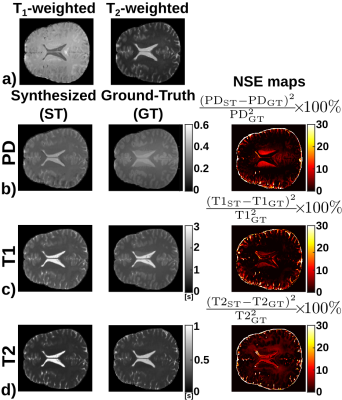 |
CNN-based synthesis of T1, T2 and PD parametric maps of the brain with a minimal input feeding
Elisa Moya-Sáez1, Óscar Peña-Nogales1, Santiago Sanz-Estébanez1, Rodrigo de Luis-Garcia1, and Carlos Alberola-López1
1Laboratorio de Procesado de Imagen, Universidad de Valladolid, Valladolid, Spain
Parametric MR maps (T1, T2 and PD) not only play a key role in quantitative imaging but they also have the capability of synthesizing any modality. However, their direct acquisition is hardly used in practice due to the need of lengthy relaxometry protocols. Synthetic MRI is a surrogate; however, no approach has been described to synthesize these maps out of a small number of customary sequences. In this work we synthesize T1, T2 and PD maps out of a T1- and a T2-weighted image using a CNN trained only with synthetic data. Our approach yields realistic maps from real data.
|
|
3807. |
Cross-Field Strength and Cross-Vendor Reliability of Quantitative MR-based Breast Density (MagDensity)
Renee Cattell1, Shenglan Chen1, Jie Ding1,2, and Chuan Huang1,3,4
1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Diagnostic Radiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong, 3Radiology, Stony Brook University, Stony Brook, NY, United States, 4Psychiatry, Stony Brook University, Stony Brook, NY, United States
Quantitative measurement of breast density is important for personalized risk assessment and frequent, longitudinal monitoring of breast density modifying drugs. Current standard of care for assessing breast density is qualitative assessment of mammogram, which involves ionizing radiation and breast compression. MRI-derived breast density (MagDensity) using fat-water decomposition has the potential to provide a sensitive and useful tool for clinicians. This study aims to prospectively determine the reliability and reproducibility of this measure across different vendors, scanner models and field strengths
|
|
3808. |
Rapid 3D sub-millimeter isotropic T1ρ mapping of the knee using motion-robust interleaved spin-lock acquisition with compressed sensing
Keita Nagawa1, Suzuki Masashi1, Masami Yoneyama2, Kaiji Inoue1, Eito Kozawa1, and Mamoru Niitsu1
1Saitama Medical University, Saitama, Japan, 2Philips Japan, Tokyo, Japan
For T1ρ quantification, a three-dimensional (3D) acquisition is desired to obtain high-resolution images. In order to achieve a rapid and robust acquisition of 3D T1 rho mapping data, we examined here the 3D sub-millimeter isotropic resolution sequences, applying them to the assessment of knee-joint. 3D isotropic resolution sequences can reduce partial-volume artifacts through the acquisition of thin continuous sections through joints. Furthermore, the isotropic source data can be used to create multiplanar reformations (MPRs). With the optimized voxel size (0.8mm3), we could obtain motion-robust high-quality isotropic images within the time constraints of a clinical exam (<10 minutes) .
|
|
3809. |
CNNs improve tissue sodium concentration accuracy in white and grey matter from stroke patients at 3T 23Na MRI
Anne Adlung1, Nadia K. Paschke1, Alena-Kathrin Schnurr1, Sherif Mohamed2, Victor Saase2, Melina Samartzi3, Marc Fatar3, Eva Neumaier-Probst2, and Lothar R. Schad1
1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 2Department of Neuroradiology, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 3Department of Neurology, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany
This study investigates the possibility to reduce 23Na MRI measurement time for stroke patients by applying CNNs and evaluates the resulting TSC accuracy in GM and WM. Three different CNN architectures were implemented and compared. The CNNs’ performance was evaluated by calculating the TSC quantification error and with a qualitative evaluation from neuroradiologists. The implementation of TSC quantification into the clinical routine might be greately facilitated by an acceleration factor of 4 for the 23Na MRI acquisition time while keeping its TSC accuracy in WM and GM.
|
|
3810. |
A joint-community effort to standardize quantitative MRI data: Updates from the BIDS extension proposal
Agah Karakuzu1,2, Gilles Hollander3,4, Stefan Appelhof5, Tibor Auer6, Mathieu Boudreau1,2, Franklin Feingold7, Ali R. Khan8, Alberto Lazari9, Christophe Phillips10, Nikola Stikov1,2, and Kirstie Whitaker11,12
1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Heart Institute, Montreal, QC, Canada, 3Laboratory for Social and Neural Systems Research (SNS Lab), Department of Economics, University of Zurich, Zürich, Switzerland, 4Spinoza Centre for Neuroimaging, Amsterdam, Netherlands, 5Center for Adaptive Rationality, Max Planck Institute for Human Development, Berlin, Germany, 6University of Surrey, Guildford, United Kingdom, 7Stanford University, Stanford, CA, United States, 8Department of Medical Biophysics, Robarts Research Institute, University of Western Ontario, London, ON, Canada, 9Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 10GIGA Institute, University of Liège, Liège, Belgium, 11Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 12The Alan Turing Institute, London, United Kingdom
The Brain Imaging Data Structure (BIDS) aims to develop a standard for organizing and describing neuroimaging data (https://bids.neuroimaging.io/). It has rapidly gained traction across neuroimaging disciplines through community-driven development of BIDS extension proposals (BEPs; http://bit.ly/bids_bep). Here were present such an extension proposal, that captures a wide range of structural MRI contrasts and parametric mapping protocols that are of interest to the broader ISMRM community (https:/bit.ly/bep001). This abstract reports the current state of the proposal and illustrates the developments made since the 2018 ISMRM virtual meeting Bringing BIDS closer to quantitative MRI.
|
|
3811.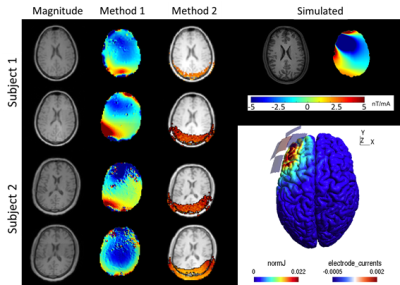 |
Magnetic Field Mapping for Transcranial Direct Current Stimulation on human brain: a preliminary study
Fábio Seiji Otsuka1, Carlos Ernesto Garrido Salmon1, Iman Ghodratitoostani2,3, Zahrasadat Vazirikangolya2,4, Renan Maatsuda5, Abhishek Datta6, and Chris Thomas6
1Inbrain Lab, Department of Physics, Faculty of Phylosophy, Sciences and Letter of Ribeirão Preto (FFCLRP), University of São Paulo (USP), Ribeirão Preto, Brazil, 2Neurocognitive Engineering Laboratory (NEL), Institute of Mathematics and Computer Sciences, University of São Paulo (USP), São Carlos, Brazil, 3Reconfigurable Computing Laboratory, Institute of Mathematics and Computer Science, University of São Paulo (USP), São Carlos, Brazil, 4Department of Neuroscience and Behavioral Sciences, Ribeirão Preto Medical School, University of São Paulo (USP), Ribeirão Preto, Brazil, 5Biomag Lab, Department of Physics, Faculty of Phylosophy, Sciences and Letter of Ribeirão Preto (FFCLRP), University of São Paulo (USP), Ribeirão Preto, Brazil, 6Soterix Medical, New York, NY, United States
Two methods to map the magnetic field distributions generated by tDCS’s electric currents using MRI were evaluated in this work. First method Stimulus/Rest Difference and second GLM. Phase images were preprocessed using motion parameters calculated by SPM from the magnitude images. To evaluate the results, simulated magnetic field distribution was calculated using simulated data for the electric current distribution. The first method showed similarities with simulated data except for one case, while the second method showed magnetic field variations only at occipital region. These results points toward the possibility of mapping these very low intensity magnetic fields using MRI.
|
|
3812.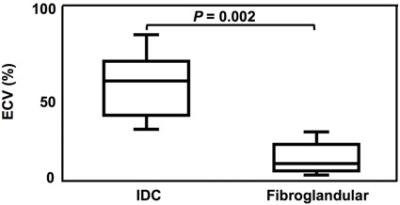 |
Evaluation of Native T1, Extracellular Volume, and apparent diffusion coefficient in Invasive Ductal Carcinoma
Ryoko Yamamori1, Akihiro Kitanaka1, Masatoshi Sakai1, Shinsuke Oie1, Naoki Ohno2, Yuki Koshino2, and Ayako Katagiri3
1Department of Radiological technology, Ishikawa Prefectural Central Hospital, Kanazawa, Japan, 2Division of Health Sciences, Institute of Sciences, Kanazawa University, Kanazawa, Japan, 3Department of Radiology, Ishikawa Prefectural Central Hospital, Kanazawa, Japan
We evaluated native T1 and extracellular volume (ECV) of the breast using modified Look-Locker inversion recovery (MOLLI) sequence and compared those between invasive ductal carcinoma (IDC) and fibroglandular tissue. Moreover, we investigated the relationship between ECV and apparent diffusion coefficient (ADC) in IDC. Our results showed that ECV of IDC was significantly higher than fibroglandular tissue, whereas there was no significant difference in native T1 between both tissues. In addition, there was no significant correlation between ECV and ADC in IDC. ECV measurement using MOLLI may provide new and more detailed information in breast cancer.
|
|
3813.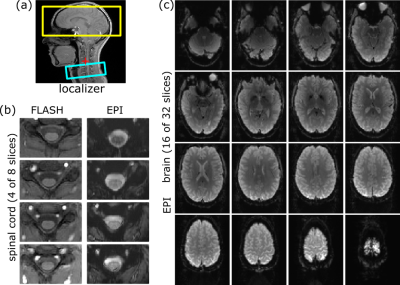 |
Optimized on-the-fly reconstruction for combined T2*-weighted imaging of the human brain and cervical spinal cord
Ying Chu1 and Jürgen Finsterbusch1
1Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Cortico-spinal functional magnetic resonance imaging (fMRI) covering slices in the brain and cervical spinal cord usually involves specific geometry (field-of-view, voxel size) and timing parameters (bandwidth) for the two volumes. This requires a time-consuming and cumbersome retrospective reconstruction after the experiment because the standard program cannot handle the different parameter settings properly, e.g., for regridding of ramp-sampled data or correction of distortions induced by Maxwell terms. Here, a reconstruction program is presented that performs an optimized reconstruction on-the-fly including the correction of receive-coil inhomogeneities without the need of a retrospective reconstruction. It could help the applicability of cortico-spinal fMRI.
|
|
3814.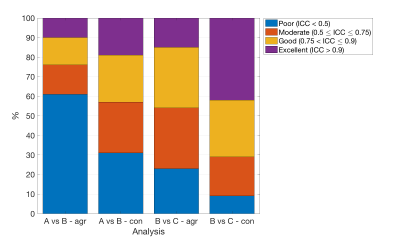 |
Blindly trusting MRI radiomics? A study on radiomic features repeatability and reproducibility with a dedicated phantom
Linda Bianchini1, João Santinha2, Nuno Loução3, Mário Figueiredo4, Francesca Botta5, Daniela Origgi5, Marta Cremonesi5, Nickolas Papanikolaou2, and Alessandro Lascialfari1
1Università degli Studi di Milano, Milan, Italy, 2Champalimaud Center for the Unknown, Lisbon, Portugal, 3Philips Healthcare, Lisbon, Portugal, 4Instituto de Telecomunicações, Instituto Superior Técnico, Lisbon, Portugal, 5Istituto Europeo di Oncologia (IEO) IRCCS, Milan, Italy
The radiomic features stability when MR scanners of different vendors or magnetic fields are involved is not known and is needed to support clinical studies. A study on the radiomic features repeatability and reproducibility was carried out with a pelvis phantom designed for radiomic purposes on three MR scanners, two of same field but different vendors and two of same vendor but different fields. The crucial results include a consistent percentage loss of features repeatability after phantom repositioning, suggesting the need for a features selection in studies involving patient’s repositioning. The limited reproducibility demands attention when dealing with multicentric studies.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance View the Poster
View the Poster Watch the Video
Watch the Video Back to Top
Back to Top