Oral - Power Pitch Session
Quantification, ML, and Tools
| Thursday Parallel 1 Live Q&A | Thursday, 13 August 2020, 14:20 - 15:05 UTC | Moderators: Daniel Gallichan & Jon Nielsen |
1037.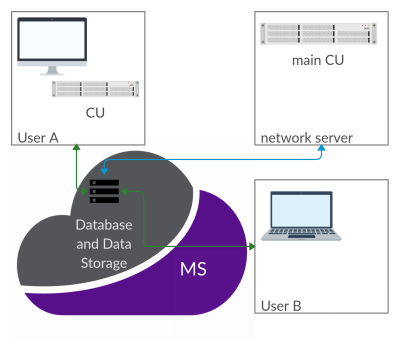 |
CAMRIE – Cloud-Accessible MRI Emulator
Eros Montin1,2, Giuseppe Carluccio1,2, Christopher Michael Collins1,2, and Riccardo Lattanzi1,2,3
1Department of Radiology, Bernard and Irene Schwartz Center for Biomedical Imaging, New York University School of Medicine, New York, NY, United States, 2Department of Radiology, Center for Advanced Imaging Innovation and Research (CAI2R), New York University School of Medicine, New York, NY, United States, 3Sackler Institute of Graduate Biomedical Sciences, New York University School of Medicine, New York, NY, United States
CAMRIE is a web-based application designed to emulate MRI experiments. It provides a numerical simulator of the Bloch equations, and enables to import electromagnetic field distributions as well as voxelized objects. A user-friendly graphic user interface guides users through the selection of predefined object geometries, with corresponding B0, B1 and gradient fields distributions. Users can then customize sequence parameters and simulate the full MRI experiment from k-space acquisition to image reconstruction. Results can be seamlessly visualized and compared across different settings. The application will be distributed via the Cloud MR portal, which allows running simulations on the cloud.
|
|
1038. |
Sycomore: an MRI simulation toolkit
Julien Lamy1 and Paulo Loureiro de Sousa1
1ICube, University of Strasbourg-CNRS, Strasbourg, France
Sycomore is an open-source MRI simulation toolkit which provides a user-friendly and consistent interface in Python for five different simulation models (Bloch simulation, three variants of EPG and the Configuration Model). Its C++ computing core, additionally helped by OpenMP, offers efficient computation of those five simulation models on desktop computers. The interactive run-time achievable for classical MRI experiments make it a valuable tool for rapid development of sequence prototypes and for teaching.
|
|
 |
1039.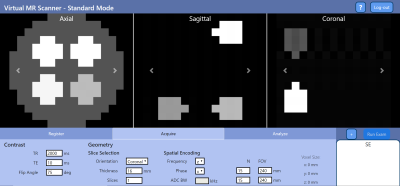 |
Virtual Scanner: MRI Experiments in a Browser
Gehua Tong1,2, Sairam Geethanath2, Keerthi Sravan Ravi1,2, Marina Manso Jimeno1,2, Enlin Qian1,2, and John Thomas Vaughan, Jr.2
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
Open-source standards for MR pulse sequences and data have been recently developed, but there is no unified platform for combining them with implemented simulation, reconstruction, and analysis tools to the best of our knowledge. We designed Virtual Scanner in order to provide a platform that allows rapid prototyping of new MR software and hardware. It also serves as a training tool for MR technicians and physicists. Two modes are provided: Standard Mode mimics MR scanner interfaces to assist training, while Advanced Mode allows customized simulation of each step in the signal chain.
|
 |
1040.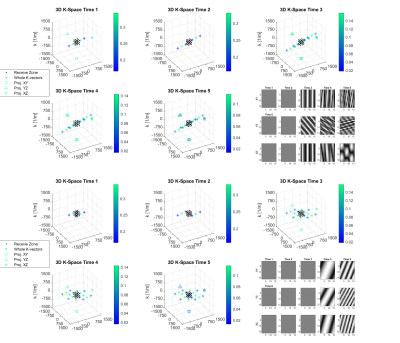 |
3D Spatially-Resolved Phase Graph
Xiang Gao1, V.G. Kiselev1, Thomas Lange1, Jürgen Hennig1, and Maxim Zaitsev1
1Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg im Breisgau, Germany A new open source recursive magnetization evolution calculation algorithm is proposed for simulating arbitrary pulse sequences efficiently and intuitively. It lifts the sequence symmetry requirements of the Extended Phase Graph and avoids intensive computations associated with direct Bloch equation simulations. The method further allows for tracking the evolution of the MR signal and corresponding k-vectors in presence of time-variant gradients with arbitrary orientations in 3D domain. To illustrate the developed technique, two simple examples are presented: spoiler design for the PRESS-based magnetic spectroscopic imaging (MRSI) and fast off-resonance calculation for dictionary building in Magnetic Resonance Fingerprinting (MRF). |
 |
1041.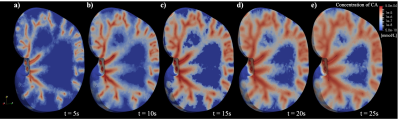 |
A dynamic digital phantom with realistic vasculature and perfusion based on MR histology
Chengyue Wu1, David A. Hormuth2, Federico Pineda3, Gregory S. Karczmar3, Robert D. Moser2,4, and Thomas E. Yankeelov1,2,5,6
1Department of Biomedical Engineering, University of Texas at Austin, Austin, TX, United States, 2Oden Institute for Computational Engineering and Sciences, University of Texas at Austin, Austin, TX, United States, 3Department of Radiology, University of Chicago, Chicago, IL, United States, 4Department of Mechanical Engineering, University of Texas at Austin, Austin, TX, United States, 5Department of Diagnostic Medicine, University of Texas at Austin, Austin, TX, United States, 6Department of Oncology, University of Texas at Austin, Austin, TX, United States
Digital phantoms are valuable tools for developing or optimizing new imaging techniques, devices, and analyses. In this contribution, we seek to develop a dynamic digital phantom which contains a detailed representation of vascular structure, tissue properties, and perfusion based on high-resolution MRI data of a rat kidney (courtesy of the Duke Center for In Vivo Microscopy). This dynamic digital phantom can be used to simulate perfusion and diffusion MRI techniques, and systematically evaluate new magnetic resonance imaging acquisition reconstruction/image processing techniques.
|
1042.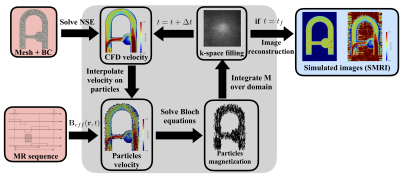 |
Numerical simulation of 4D Flow MRI
Thomas Puiseux1,2, Anou Sewonu1,3, Ramiro Moreno1,3,4, Simon Mendez2, and Franck Nicoud2
Video Permission Withheld
1SPIN UP, Toulouse, France, 2IMAG, Univ. Montpellier, CNRS, Montpellier, France, 3I2MC, INSERM U1048, Toulouse, France, 4ALARA Expertise, Strasbourg, France
The present study proposes a novel approach to efficiently simulate 4D Flow MRI acquisitions in realistic complex flow conditions. Navier-Stokes and Bloch equations are simultaneously solved with Eulerian-Lagrangian coupling. A semi-analytic solution for the Bloch equation as well as a periodic particle re-injection strategy are implemented to reduce the computational cost. The Bloch solver and the velocity reconstruction pipeline were first validated in a steady flow configuration. The coupled 4D Flow MRI simulation procedure was validated in a complex pulsatile flow phantom cardiovascular-typical experiment. Besides, we compared simulated MR velocity data with experimental 4D Flow MRI measurements.
|
|
 |
1043.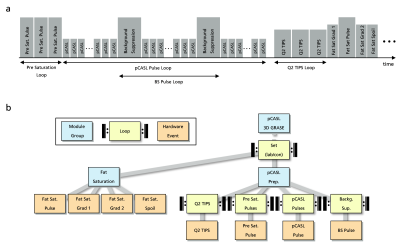 |
Event-Based Traversing of Hierarchical Sequences Allows Real-Time Execution and Arbitrary Looping in a Scanner-Independent MRI Framework
Daniel Christopher Hoinkiss1, Cristoffer Cordes1, Simon Konstandin1, and Matthias Günther1,2
1MR Physics, Fraunhofer MEVIS, Bremen, Germany, 2MR-Imaging & Spectroscopy, Faculty 01 (Physics/Electrical Engineering), University of Bremen, Bremen, Germany
MR sequence development is either based on complex, platform-specific solutions or restricted by fixed sequence structures together with a strict hierarchical implementation of loops. This abstract introduces event-based traversing of gammaSTAR sequences together with a buffered real-time execution at the scanner. The concept provides easy implementation of arbitrary, interleaved loop structures as well as memory efficient, real-time capable sequence execution in a vendor-agnostic environment. It is demonstrated for interleaved loop structures in a pCASL 3D GRASE sequence for brain perfusion imaging.
|
1044.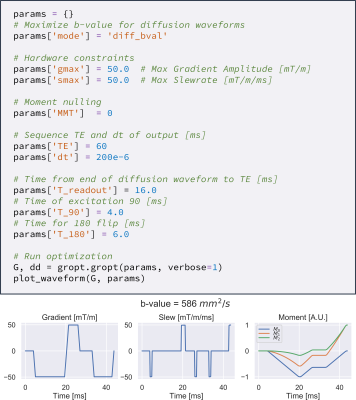 |
Gradient Optimization (GrOpt) Toolbox: A Software Package for Fast Gradient Waveform Design
Michael Loecher1,2, Matthew Middione1,2, and Daniel B Ennis1,2,3,4
1Radiology, Stanford, Palo Alto, CA, United States, 2Radiology, Veterans Administration Health Care System, Palo Alto, CA, United States, 3Cardiovascular Institute, Stanford, Palo Alto, CA, United States, 4Center for Artificial Intelligence in Medicine & Imaging, Stanford, Palo Alto, CA, United States
Objective: To introduce and demonstrate a software library for time-optimal gradient waveform optimization for a wide range of applications. The software allows for direct just-in-time gradient waveform design on scanner hardware for multiple vendors. The software is tested over a range of constraints and acquisition types for which compute times are on the order of (1-100ms). The sequences are also implemented on two different vendor scanners, demonstrating the interoperability of the method.
|
|
 |
1045.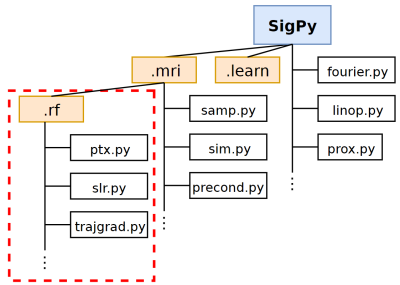 |
SigPy.RF: Comprehensive Open-Source RF Pulse Design Tools for Reproducible Research
Jonathan B Martin1, Frank Ong2, Jun Ma1, Jonathan I Tamir3,4, Michael Lustig3, and William A Grissom1
1Biomedical Engineering, Vanderbilt University, Nashville, TN, United States, 2Electrical Engineering, Stanford University, Stanford, CA, United States, 3Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 4Electrical and Computer Engineering, UT Austin, Austin, TX, United States
We present SigPy.RF, an extensive set of open-source, Python-based tools for MRI RF pulse design. This toolbox extends the SigPy Python software package and leverages SigPy’s existing capabilities for GPU computation, iterative optimization, and powerful abstractions for linear operators, proximal operators, and applications. Tools are available for all steps of the excitation design process including trajectory/gradient design, pulse design, and simulation. Our implemented functions for pulse design include advanced SLR, multiband, adiabatic, optimal control, B$$$_1^+$$$-selective and small-tip pTx designers. SigPy.RF pulse designs were validated in simulations and a pTx experiment.
|
 |
1046.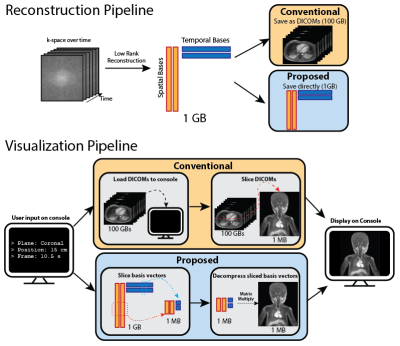 |
A Reconstruction Compatible, Fast and Memory Efficient Visualization Framework for Large-scale Volumetric Dynamic MRI
Cedric Yue Sik Kin1, Frank Ong2, Jonathan I Tamir3,4, Michael Lustig3, John M Pauly2, and Shreyas S Vasanawala1
1Radiology, Stanford University, Stanford, CA, United States, 2Electrical Engineering, Stanford University, Stanford, CA, United States, 3Electrical and Computer Sciences, UC Berkeley, Berkeley, CA, United States, 4Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States
We addressed the speed and memory shortcomings of conventional visualization consoles when processing high-dimensional MRI datasets by proposing a novel approach that leverages compressed representations of such datasets. We considered low rank reconstructions and operated on them directly for visualization, unlike traditional viewers which load entire uncompressed image datasets. We built a web viewer that utilizes this approach to demonstrate real time reformatting and slicing. We were able to achieve more than 15x reduction in both memory usage and loading times.
|
1047.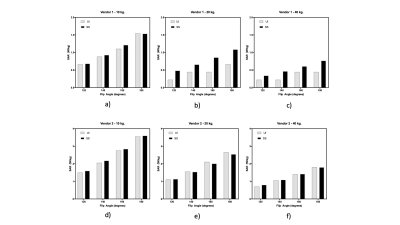 |
Specific Absorption Rate prediction for open source pulse sequence programming
Sairam Geethanath1, Jon-Fredrik Nielsen2, Douglas C Noll2, and John Thomas Vaughan Jr.1
1Columbia MR Research Center, Columbia University, New York, NY, United States, 2University of Michigan, Ann Arbor, MI, United States
Flexibility in designing custom pulse sequences has a direct impact on the development of diverse MR techniques and strategies. However, it is important to be cognizant of the MR safety risks that are associated with such custom sequences. We develop and evaluate an open source software package to predict global Specific Absorption Rate for Pulseq and TOPPE based sequences. We compare the resulting predictions with scanner reported SAR values for different flip angles and three sample weights, on two major MR vendor platforms. The predictions correlate highly with the scanner reported values: R2 = 0.87 (vendor 1); 0.99 (vendor 2).
|
|
1048.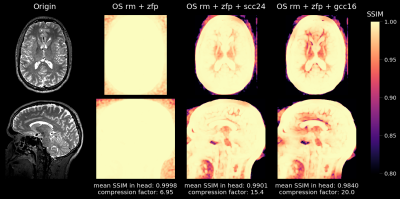 |
MRI Raw Data Compression for long-term storage in large-scale population imaging
Philipp Ehses1, Marten Veldmann1, Yiming Dong1, and Tony Stöcker1
1German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany
In the overwhelming majority of MRI studies, only the reconstructed images are stored and the raw data that was used during the reconstruction process is lost. However, routine raw data storage would potentially allow imaging studies that are conducted now to benefit from future improved image reconstruction techniques. Unfortunately, the raw data storage requirements are often prohibitive, especially in large-scale population studies. We developed a flexible software tool that achieves high lossy compression of MRI raw data and is able to decompress the data back to the vendor-specific format to allow for retrospective reconstruction using the vendor's reconstruction pipeline.
|
|
 |
1049. |
Chemical sHift bAsed pRospectIve k-Space anonyMizAtion (CHARISMA)
Hendrik Mattern1, Martin Knoll1, Falk Lüsebrink 1,2, and Oliver Speck1,3,4,5
1Biomedical Magnetic Resonance, Otto-von-Guericke University, Magdeburg, Germany, 2Medicine and Digitalization, Otto-von-Guericke University, Magdeburg, Germany, 3German Center for Neurodegenerative Disease, Magdeburg, Germany, 4Center for Behavioral Brain Sciences, Magdeburg, Germany, 5Leibniz Institute for Neurobiology, Magdeburg, Germany One key element of open science is to make all data publicly available. In case of neuroscience, reconstructed images can be defaced to prevent data privacy violations, but no strategy to anonymize raw data has been presented to our best knowledge. Here, chemical shift based prospective k-Space anonymization is presented. The subject wears an oil-filled mask which is superimposed onto the subject’s skin due to chemical shift. This low-cost solution (<15€) is easy to build and applicable for sequences with sufficient chemical shift in the A-P direction. |
 |
1050.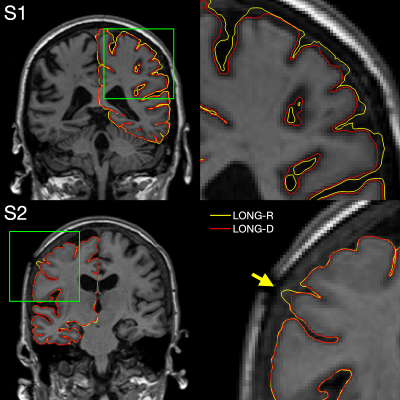 |
Longitudinal FreeSurfer with non-linear subject-specific template improves sensitivity to cortical thinning
Malte Hoffmann1,2, David Salat1,2, Martin Reuter*1,2,3, and Bruce Fischl*1,2,4
1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3German Center for Neurodegenerative Diseases, Bonn, Germany, 4Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States
Longitudinal FreeSurfer creates a within-subject template by rigidly registering and median-filtering longitudinal timepoints (TP). Information common to all TPs is extracted from the template for unbiased TP initialization, resulting in substantial improvements over cross-sectional processing. However, this approach is not optimal in the presence of severe atrophy or other large-scale anatomical change, which causes voxels to be filtered across tissue classes. We address this problem by introducing an enhanced longitudinal stream that deforms each TP using non-linear registration to construct the template. We demonstrate considerable increases in sensitivity to cortical thinning, without affecting test-retest reliability.
|
1051.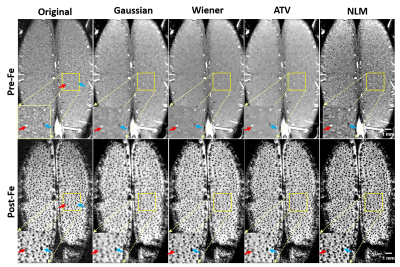 |
Developing a Novel and Robust Preprocessing Pipeline for Intensity-Based High-Resolution Magnetic Resonance Angiogram
Wei Zhu1, Yi Zhang1, Xiao-Hong Zhu1, and Wei Chen1
1Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, United States
In-vivo high-resolution imaging of cerebral blood vessels is critical for brain functional research and clinical diagnosis. Despite well-developed magnetic resonance angiogram (MRA) techniques, a simple, robust preprocessing procedure has yet to be established. Thus, we propose a preprocessing pipeline that includes zero-fill interpolation, intensity non-uniformity correction, image denoising, vessel enhancement and segmentation. Specifically, we found that the most effective and robust denoising method is anisotropic total variation (ATV). By adopting and optimizing an improved 3D Hessian based tubular and spherical enhancement filter and a region-based level-set image segmentation method, we can automate the preprocessing of intensity-based MRAs with high fidelity.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video Back to Top
Back to Top