Farshid Sepehrband1, Giuseppe Barisano2, Hyun-Joon Yang2, Jeiran Choupan2, and Arthur W Toga2
1Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, USC, Los Angeles, CA, United States, 2USC, Los Angeles, CA, United States
1Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, USC, Los Angeles, CA, United States, 2USC, Los Angeles, CA, United States
Combining T1w and FLAIR enables PVS mapping of clinical data and could also improve WMH mapping. In particular, we showed that PVS can be mapped in clinical data.
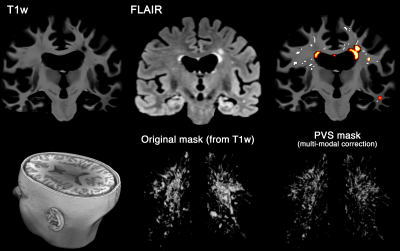
Figure 1. Multi-modal PVS segmentation of clinical data. PVS were mapped from T1w images and then white matter hyperintensities, which was falsely segmented as PVS, were excluded using FLAIR images. PVS segmentation was done using an automated and highly reliable quantification technique [3], based on non-local filtering, Frangi filtering and optimized mask identification. For each segmented voxel in T1w, corresponding FLAIR voxels was checked, and if the FLAIR voxel value was a white matter hyperintensity, then the voxel was excluded from the final PVS mask.
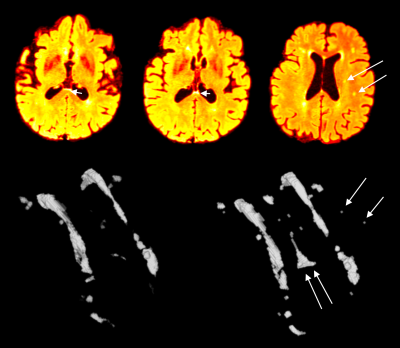
Figure 2. Multi-modal CNN-based WMH segmentation. WMH segmentation when compared when an algorithmic based approach was used[4](left 3D segmented WMH) versus when a multi-modal CNN-based approach was used. Note the increased sensitivity to the small WMHs, or WMH at the ventricle boundary which is often challenging to detect.