Digital Posters
Machine Learning for Image Analysis
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 19:00 - 20:00 |
 |
2397.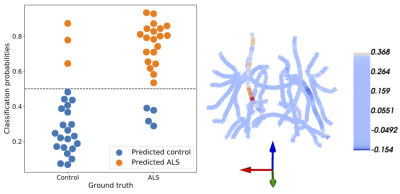 |
Multidimensional analysis and detection of informative features in diffusion MRI measurements of human white matter
Adam C Richie-Halford1, Jason Yeatman2, Noah Simon3, and Ariel Rokem4
1eScienceInstitute, University of Washington, Seattle, WA, United States, 2Graduate School of Education and Division of Developmental and Behavioral Pediatrics, Stanford University, Stanford, CA, United States, 3Department of Biostatistics, University of Washington, Seattle, WA, United States, 4Department of Psychology, University of Washington, Seattle, WA, United States
We present a novel method for the analysis of diffusion MRI tractometry data based on the sparse group lasso. It capitalizes on natural anatomical grouping of diffusion metrics, providing both accurate prediction of phenotypic information and results that are readily interpretable. We show the effectiveness of this approach in two settings. In a classification setting, patients with amyotrophic lateral sclerosis (ALS) are accurately distinguished from matched controls and SGL automatically identifies known anatomical correlates of ALS. In a regression setting, we accurately predict “brain age” in two previous dMRI studies. We demonstrate that our approach is both accurate and interpretable.
|
||
2398.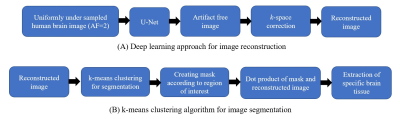 |
A Deep k-means Based Tissue Extraction from Reconstructed Human Brain MR Image
Madiha Arshad1, Mahmood Qureshi1, Omair Inam1, and Hammad Omer1
1Medical Image Processing Research Group (MIPRG), Department of Electrical and Computer Engineering, COMSATS University, Islamabad, Pakistan
Fast and accurate tissue extraction of human brain is an ongoing challenge. Two principal factors make this task difficult:(1) quality of the reconstructed images, (2) accuracy and availability of the segmentation masks. In this proposed method, firstly, a supervised deep learning framework is used for the reconstruction of solution image from the acquired uniformly under-sampled human brain data. Later, an unsupervised clustering approach i.e. k-means is used for the extraction of specific tissue from the reconstructed image. Experimental results show a successful extraction of cerebrospinal fluid (CSF), white matter and grey matter from the human brain image.
|
|||
2399.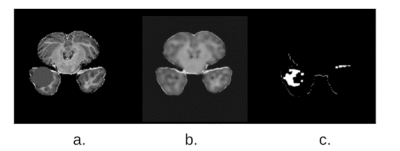 |
Unsupervised reconstruction based anomaly detection using a Variational Auto Encoder
Soumick Chatterjee1,2,3, Alessandro Sciarra1,4, Max Dünnwald3,4, Shubham Kumar Agrawal3, Pavan Tummala3, Disha Setlur3, Aman Kalra3, Aishwarya Jauhari3, Steffen Oeltze-Jafra4,5,6, Oliver Speck1,5,6,7, and Andreas Nürnberger2,3,6
1Department of Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4MedDigit, Department of Neurology, Medical Faculty, University Hopspital, Magdeburg, Germany, 5German Centre for Neurodegenerative Diseases, Magdeburg, Germany, 6Center for Behavioral Brain Sciences, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
While commonly used approach for disease localization, we propose an approach to detect anomalies by differentiating them from reliable models of anatomies without pathologies. The method is based on a Variational Auto Encoder to learn the anomaly free distribution of the anatomy and a novel image subtraction approach to obtain pixel-precise segmentation of the anomalous regions. The proposed model has been trained with the MOOD dataset. Evaluation is done on BraTS 2019 dataset and a subset of the MOOD, which contain anomalies to be detected by the model.
|
|||
2400.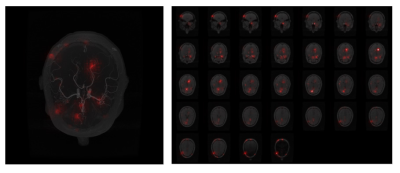 |
Interpretability Techniques for Deep Learning based Segmentation Models
Soumick Chatterjee1,2,3, Arnab Das3, Chirag Mandal3, Budhaditya Mukhopadhyay3, Manish Vipinraj3, Aniruddh Shukla3, Oliver Speck1,4,5,6, and Andreas Nürnberger2,3,6
1Department of Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4German Centre for Neurodegenerative Diseases, Magdeburg, Germany, 5Leibniz Institute for Neurobiology, Magdeburg, Germany, 6Center for Behavioral Brain Sciences, Magdeburg, Germany
In medical image analysis, it is desirable to decipher the black-box nature of Deep Learning models in order to build confidence in clinicians while using such methods. Interpretability techniques can help understand the model’s reasonings, e.g. by showcasing the anatomical areas the network focuses on. While most of the available interpretability techniques work with classification models, this work presents various interpretability techniques for segmentation models and shows experiments on a vessel segmentation model. In particular, we focus on input attributions and layer attribution methods which give insights on the critical features of the image identified by the model.
|
|||
2401.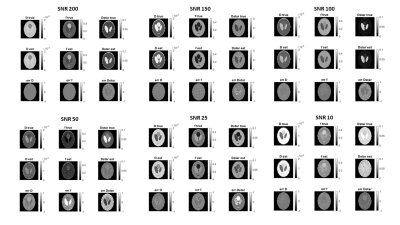 |
A Supervised Artificial Neural Network Approach with Standardized Targets for IVIM Maps Computation
Alfonso Mastropietro1, Daniele Procissi2, Elisa Scalco1, Giovanna Rizzo1, and Nicola Bertolino2
1Istituto di Tecnologie Biomediche, Consiglio Nazionale delle Ricerche, Segrate, Italy, 2Radiology, Northwestern University, Chicago, IL, United States
Fitting the IVIM bi-exponential model is challenging especially at low SNRs and time consuming. In this work we propose a supervised artificial neural network approach to obtain reliable parameters estimation as demonstrated in both simulated data and real acquisition. The proposed approach is promising and can outperform, in specific conditions, other state-of-the-art fitting methods.
|
|||
2402. |
Task Performance or Artifact Reduction? Evaluating the Number of Channels and Dropout based on Signal Detection on a U-Net with SSIM Loss
Rachel E Roca1, Joshua D Herman1, Alexandra G O'Neill1, Sajan G Lingala2, and Angel R Pineda1
1Mathematics Department, Manhattan College, Riverdale, NY, United States, 2Roy J. Carver Department of Biomedical Engineering, University of Iowa, Iowa City, IA, United States
The changes in image quality caused by varying the parameters and architecture of neural networks are difficult to predict. It is important to have an objective way to measure the image quality of these images. We propose using a task-based method based on detection of a signal by human and ideal observers. We found that choosing the number of channels and amount of dropout of a U-Net based on the simple task we considered might lead to images with artifacts which are not acceptable. Task-based optimization may not align with artifact minimization.
|
|||
2403.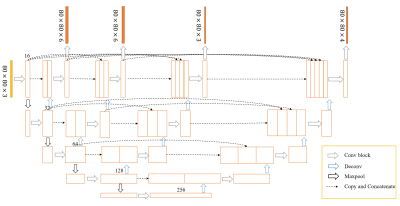 |
Deep Learning for Automated Segmentation of Brain Nuclei on Quantitative Susceptibility Mapping
Yida Wang1, Naying He2, Yan Li2, Yi Duan1, Ewart Mark Haacke2,3, Fuhua Yan2, and Guang Yang1
1East China Normal University, Shanghai Key Laboratory of Magnetic Resonance, Shanghai, China, 2Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 3Department of Biomedical Engineering, Wayne State University, Detroit, MI, United States
We proposed a deep learning (DL) method to automatically segment brain nuclei including caudate nucleus, globus pallidus, putamen, red nucleus, and substantia nigra on Quantitative Susceptibility Mapping (QSM) data. Due to the large differences of shape and size of brain nuclei, the output branches at different semantic levels in U-net++ model were designed to simultaneously output different brain nuclei. Deep supervision was applied for improving segmentation performance. The segmentation results showed the mean Dice coefficients for the five nuclei achieved a value above 0.8 in validation dataset and the trained network could accurately segment brain nuclei regions on QSM images.
|
|||
2404. |
Task-Based Assessment for Neural Networks: Evaluating Undersampled MRI Reconstructions based on Signal Detection
Joshua D Herman1, Rachel E Roca1, Alexandra G O'Neill1, Sajan G Lingala2, and Angel R Pineda1
1Mathematics Department, Manhattan College, Riverdale, NY, United States, 2Roy J. Carver Department of Biomedical Engineering, University of Iowa, Iowa City, IA, United States
Artifacts from neural network reconstructions are difficult to characterize. It is important to assess the image quality in terms of the task for which the images will be used. In this work, we evaluated the effect of undersampling on detection of signals in images reconstructed with a neural network by both human and ideal observers. We compared these results to standard metrics (SSIM and NRMSE). Our results suggest that the undersampling level chosen by SSIM, NRMSE and ideal observer would likely be different than that of a human observer on a detection task for a small signal.
|
|||
2405.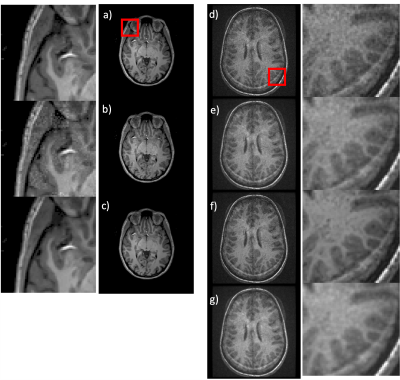 |
MRI denoising using native noise
Sairam Geethanath1, Pavan Poojar1, Keerthi Sravan Ravi1, and Godwin Ogbole2
1Columbia University, New York, NY, United States, 2Department of Radiology, University College Hospital(UCH) Ibadan, Ibadan, Nigeria
The benefits of deep learning (DL) based denoising of MR images include reduced acquisition time and improved image quality at low field strength. However, simulating noisy images require biophysical models that are field and acquisition dependent. Scaling these simulations is complex and computationally intensive. In this work, we instead leverage the native noise of the data, dubbed “native noise denoising network” (NNDnet). We applied NNDnet to three different MR data types and computed the peak signal-to-noise ratio (> 38dB) for training performance and image entropy (> 4.25) for testing performance in the absence of a reference image.
|
|||
 |
2406.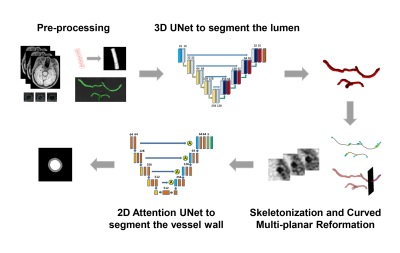 |
A fully automated framework for intracranial vessel wall segmentation based on 3D black-blood MRI
Jiaqi Dou1, Hao Liu1, Qiang Zhang1, Dongye Li2, Yuze Li1, Dongxiang Xu3, and Huijun Chen1
1Center for Biomedical Imaging Research, School of Medicine, Tsinghua University, Beijing, China, 2Department of Radiology, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, China, 3Department of Radiology, University of Washington, Seattle, WA, United States
Intracranial atherosclerosis is a major cause of stroke worldwide. Vessel wall quantitative measurement is an essential tool for plaque analysis, while manual vessel wall segmentation is time-consuming and costly. In this study, we proposed a fully automated vessel wall segmentation framework for intracranial arteries using only 3D black-blood MRI, in which 3D lumen segmentation and skeletonization were applied to locate the arteries of interest for further 2D vessel wall segmentation. It achieved high segmentation performance for both normal (DICE=0.941) and stenotic (DICE=0.922) vessel wall and provided a promising tool for quantitative intracranial atherosclerosis analysis in large population studies.
|
||
2407.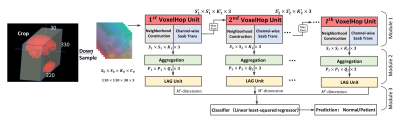 |
Successive Subspace Learning for ALS Disease Classification Using T2-weighted MRI
Xiaofeng Liu1, Fangxu Xing1, Chao Yang2, C.-C. Jay Kuo3, Suma Babu4, Georges El Fakhri1, Thomas Jenkins5, and Jonghye Woo1
1Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States, 2Facebook AI, Boston, MA, United States, 3Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 4Sean M Healey & AMG Center for ALS, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, BOSTON, MA, United States, 5Sheffield Institute for Translational Neuroscience, University of Sheffield, Sheffield, United Kingdom
A challenge in Amyotrophic Lateral Sclerosis (ALS) research and clinical practice is to detect the disease early to ensure patients have access to therapeutic trials in a timely manner. To this end, we present a successive subspace learning model for accurate classification of ALS from T2-weighted MRI. Compared with popular CNNs, our method has modular structures with fewer parameters, so is well-suited to small dataset size and 3D data. Our approach, using 20 controls and 26 patients, achieved an accuracy of 93.48% in differentiating patients from controls, which has a potential to help aid clinicians in the decision-making process.
|
|||
2408.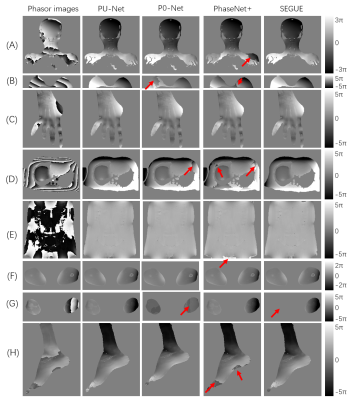 |
PU-NET: A robust phase unwrapping method for magnetic resonance imaging based on deep learning
Hongyu Zhou1, Chuanli Cheng1, Xin Liu1, Hairong Zheng1, and Chao Zou1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
This work proposed a robust MR phase unwrapping method based on a deep-learning method. Through comparisons of MR images over the entire body, the model showed promising performances in both unwrapping errors and computation times. Therefore, it has promise in applications that use MR phase information.
|
|||
2409.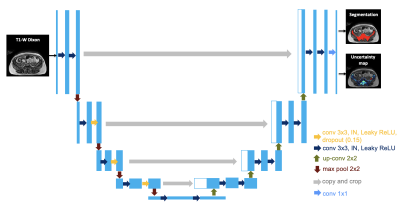 |
Using uncertainty estimation to increase the robustness of bone marrow segmentation in T1-weighted Dixon MRI for multiple myeloma
Renyang Gu1, Michela Antonelli1, Pritesh Mehta 2, Ashik Amlani 3, Adrian Green3, Radhouene Neji 4, Sebastien Ourselin1, Isabel Dregely1, and Vicky Goh1
1School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 2Biomedical Engineering and Medical Physics, University College London, London, United Kingdom, 3Radiology, Guy’s and St Thomas’ Hospitals, London, United Kingdom, 4Siemens Healthcare Limited, Frimley, United Kingdom
Reliable skeletal segmentation of T1-weighted Dixon MRI is a first step towards measuring marrow fat-fraction as a surrogate metric for early marrow infiltration. We proposed an uncertainty-aware 2D U-Net (uU-Net) to reduce the impact of noisy ground-truth labels on segmentation accuracy. Five-fold cross-validation on a dataset of 30 myeloma patients provided a mean ± SD Dice coefficient of 0.74 ± 0.03 (vs. 0.73 ± 0.04, U-Net) and 0.63 ± 0.03 (vs 0.62 ± 0.04, U-Net) for pelvic and abdominal stations, respectively. Of clinical importance, improved segmentation of the ilium and vertebrae were achieved.
|
|||
2410.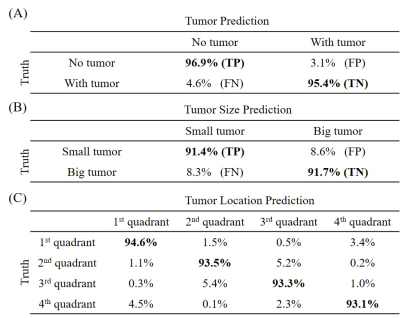 |
Deep Learning Pathology Detection from Extremely Sparse K-Space Data
Linfang Xiao1,2, Yilong Liu1,2, Zheyuan Yi1,2,3, Yujiao Zhao1,2, Peiheng Zeng1,2, Alex T.L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
Traditional MRI diagnosis consists of image reconstruction from k-space data and pathology identification in the image domain. In this study, we propose a strategy of direct pathology detection from extremely sparse MR k-space data through deep learning. This approach bypasses the traditional MR image reconstruction procedure prior to pathology diagnosis and provides an extremely rapid and potentially powerful tool for automatic pathology screening. Our results demonstrate that this new approach can detect brain tumors and classify their sizes and locations directly from single spiral k-space data with high sensitivity and specificity.
|
|||
2411.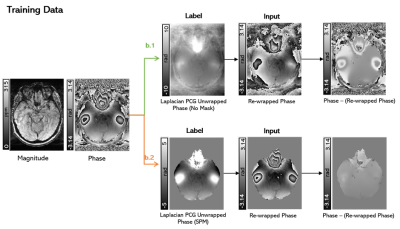 |
Development of a Deep Learning MRI Phase Unwrapping Technique Using an Exact Unwrap-Rewrap Training Model
Rita Kharboush1, Anita Karsa1, Barbara Dymerska1, and Karin Shmueli1
1Medical Physics and Biomedical Engineering, University College London, London, United Kingdom
No existing phase unwrapping technique achieves completely accurate unwrapping. Therefore, we trained a convolutional neural network for phase unwrapping on (flipped and scaled) brain images from 12 healthy volunteers. An exact model of phase unwrapping was used: ground-truth (label) phase images (unwrapped with an iterative Laplacian Preconditioned Conjugate Gradient technique) were rewrapped (projected into the 2π range) to provide input images. This novel model can be used to train any neural network. Networks trained using masked (and unmasked) images showed unwrapping performance similar to state-of-the-art SEGUE phase unwrapping on test brain images and showed some generalisation to pelvic images.
|
|||
2412.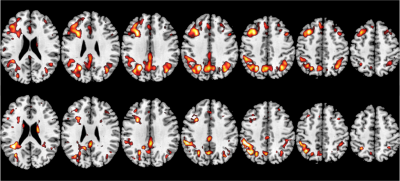 |
Improving ASL MRI Sensitivity for Clinical Applications Using Transfer Learning-based Deep Learning
Danfeng Xie1, Yiran Li1, and Ze Wang1
1Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States
This study represents the first effort to apply transfer learning of Deep learning-based ASL denoising (DLASL) method on clinical ASL data. Pre-trained with young healthy subjects’ data, DLASL method showed improved Contrast-to-Noise Ratio (CNR) and Signal-to-Noise Ratio (SNR) and higher sensitivity for detecting the AD related hypoperfusion patterns compared with the conventional method. Experimental results demonstrated the high transfer capability of DLASL for clinical studies.
|
|||
2413.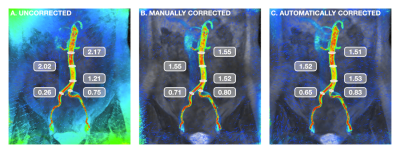 |
Fully-Automated Deep Learning-Based Background Phase Error Correction for Abdominopelvic 4D Flow MRI
Sophie You1, Evan M. Masutani1, Joy Liau2, Marcus T. Alley3, Shreyas S. Vasanawala3, and Albert Hsiao2
1School of Medicine, University of California, San Diego, La Jolla, CA, United States, 2Department of Radiology, University of California, San Diego, La Jolla, CA, United States, 3Department of Radiology, Stanford University School of Medicine, Stanford, CA, United States
4D Flow MRI is valuable for the evaluation of cardiovascular disease, but abdominal applications are currently limited by the need for background phase error correction. We propose an automated deep learning-based method that utilizes a multichannel 3D convolutional neural network (CNN) to produce corrected velocity fields. Comparisons of arterial and venous flow, as well as flow before and after bifurcation of major abdominal vessels, show improved flow continuity with greater agreement after automated correction. Results of automated corrections are comparable to manual corrections. CNN-based corrections may improve reliability of flow measurements from 4D Flow MRI.
|
|||
2414.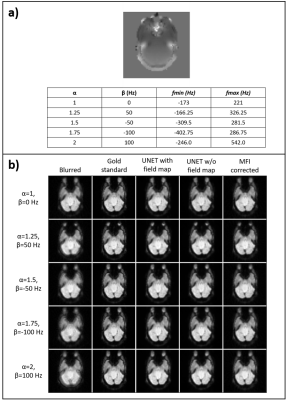 |
Deblurring of spiral fMRI images using deep learning
Marina Manso Jimeno1,2, John Thomas Vaughan Jr.1,2, and Sairam Geethanath2
1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States
fMRI acquisitions benefit from spiral trajectories; however, their use is commonly restricted due to off-resonance blurring artifacts. This work presents a deep-learning-based model for spiral deblurring in inhomogeneous fields. Training of the model utilized blurred simulated images from interleaved EPI data with various degrees of off-resonance. We investigated the effect of using the field map during training and compared correction performance with the MFI technique. Quantitative validation results demonstrated that the proposed method outperforms MFI for all inhomogeneity scenarios with SSIM>0.97, pSNR>35 dB, and HFEN<0.17. Filter visualization suggests blur learning and mitigation as expected.
|
|||
2415.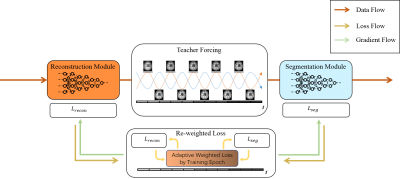 |
Multi-task MR imaging with deep learning
Kehan Qi1, Yu Gong1,2, Haoyun Liang1, Xin Liu1, Hairong Zheng1, and Shanshan Wang1
1Paul C Lauterbur Research Center, Shenzhen Inst. of Advanced Technology, shenzhen, China, 2Northeastern University, Shenyang, China
Noises, artifacts, and loss of information caused by the MR reconstruction may compromise the final performance of the downstream applications such as image segmentation. In this study, we develop a re-weighted multi-task deep learning method to learn prior knowledge from the existing big dataset and then utilize them to assist simultaneous MR reconstruction and segmentation from under-sampled k-space data. It integrates the reconstruction with segmentation and produces both promising reconstructed images and accurate segmentation results. This work shows a new way for the direct image analysis from k-space data with deep learning.
|
|||
2416.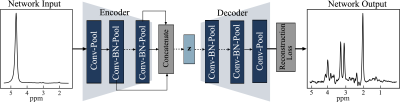 |
Quantification of Unsuppressed Water Spectrum using Autoencoder with Feature Fusion
Marcia Sahaya Louis1,2, Eduardo Coello2, Huijun Liao2, Ajay Joshi1, and Alexander P Lin2
1Boston University, Boston, MA, United States, 2Brigham and Women's hospital, Boston, MA, United States
Recent years have witnessed novel applications of machine learning in radiology. Developing robust machine learning based methods for removing spectral artifacts and reconstructing the intact metabolite spectrum is an open challenge in MR spectroscopy (MRS). We had shown autoencoder models reconstruct metabolite spectrum from unsuppressed water spectrum for short TE with relatively high SNR. In this work we presents an autoencoder model with feature fusion method to extract the shallow and deep features from a water unsuppressed 1H MR spectrum. The model learns to map the extracted feature to a latent code and reconstruct the intact metabolite spectrum
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.