Oral
Machine Learning for Image Analysis
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 18:00 - 20:00 | Moderators: Sila Kurugol & Yunyan Zhang |
0387.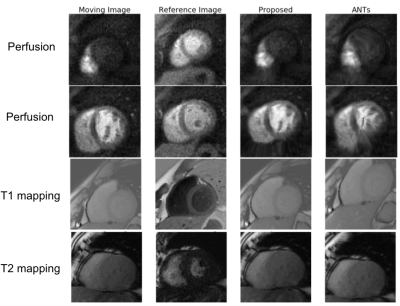 |
Deep-Learning-Based Motion Correction For Quantitative Cardiac MRI
Alfredo De Goyeneche1, Shuyu Tang1, Nii Okai Addy1, Bob Hu1, William Overall1, and Juan Santos1
1HeartVista, Inc., Los Altos, CA, United States
We developed a deep-learning-based approach for motion correction in quantitative cardiac MRI, including perfusion, T1 mapping, and T2 mapping. The proposed approach consists of a segmentation network and a registration network. The segmentation network was trained using 2D short-axis images for each of the three sequences, while the same registration network was shared between all three sequences. The proposed approach was faster and more accurate than a popular traditional registration method. Our work is beneficial for building a faster and more robust automated processing pipeline to obtain CMR parametric maps.
|
||
 |
0388.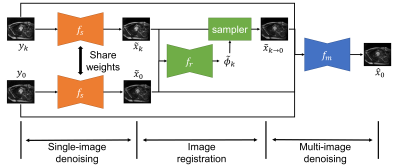 |
Self-supervised Cardiac MRI Denoising with Deep Learning
Junshen Xu1 and Elfar Adalsteinsson1
1Massachusetts Institute of Technology, Cambridge, MA, United States
Image denoising is of great importance for medical imaging system, since it can improve image quality for disease diagnosis and further image processing. In cardiac MRI, images are acquired at different time frames to capture the cardiac dynamic. The correlation among different time frames makes it possible to improve denoising results with information from other time frames. In this work, we propose a self-supervised deep learning framework for cardiac MRI denoising. Evaluation on in vivo data with different noise statistics shows that our method has comparable or even better performance than other state-of-the-art unsupervised or self-supervised denoising methods.
|
|
0389.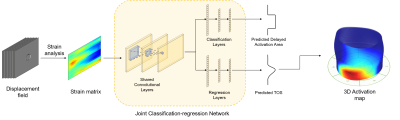 |
Multi-task Deep Learning for Late-activation Detection of Left Ventricular Myocardium
Jiarui Xing1, Sona Ghadimi2, Mohammad Abdishektaei2, Kenneth C Bilchick3, Frederick H Epstein2, and Miaomiao Zhang1
1Electrical and Computer Engineering, University of Virginia, Charlottesville, VA, United States, 2Department of Biomedical Engineering, University of VIrginia, Charlottesville, VA, United States, 3School of Medicine, University of Virginia, Charlottesville, VA, United States
Implantation of the left ventricular pacing lead at the area with delayed activation is critical to Cardiac Resynchronization Therapy (CRT) response. Current approaches of detecting late-activated regions of left ventricles (LV) are slow with unsatisfied accuracy, particularly in cases where scar tissues exist in the patient’s heart. This work presents a multi-task deep learning algorithm to automatically identify late-activated regions of LV, as well as estimating the Time to the Onset of circumferential Shortening (TOS) using spatio-temporal cardiac DENSE MR images. Experimental results show that our algorithm provides ultra-fast identification of late-activated regions and estimated TOS with increased accuracy.
|
||
 |
0390.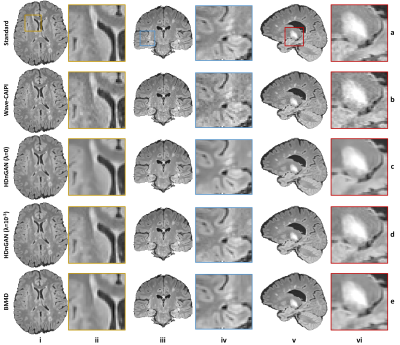 |
HDnGAN: High-fidelity ultrafast volumetric brain MRI using a hybrid denoising generative adversarial network
Ziyu Li1, Qiyuan Tian2, Chanon Ngamsombat2,3, Samuel Cartmell4, John Conklin2,4, Augusto Lio M. Gonçalves Filho2,4, Wei-Ching Lo5, Guangzhi Wang1, Kui Ying6, Kawin Setsompop7,8, Qiuyun Fan2, Berkin Bilgic2, Stephen Cauley2,
and Susie Y Huang2,4
1Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, United States, 3Department of Radiology, Siriraj Hospital, Mahidol University, Bangkok, Thailand, 4Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 5Siemens Medical Solutions, Boston, MA, United States, 6Department of Engineering Physics, Tsinghua University, Beijing, China, 7Department of Radiology, Stanford University, Stanford, CA, United States, 8Department of Electrical Engineering, Stanford University, Stanford, CA, United States
Highly accelerated high-resolution volumetric brain MRI is intrinsically noisy. A hybrid generative adversarial network (GAN) for denoising (entitled HDnGAN) consisting of a 3D generator and a 2D discriminator was proposed to improve the SNR of highly accelerated images while preserving realistic textures. The novel architecture benefits from improved image synthesis performance and increased training samples for training the discriminator. HDnGAN's efficacy is demonstrated on 3D standard and Wave-CAIPI T2-weighted FLAIR data acquired in 33 multiple sclerosis patients. Generated images are similar to standard FLAIR images and superior to Wave-CAIPI and BM4D-denoised images in quantitative evaluation and assessment by neuroradiologists.
|
|
0391.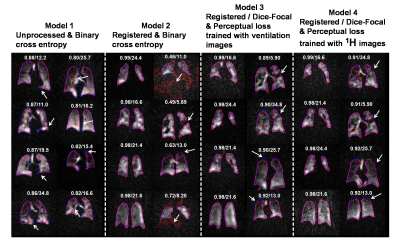 |
Deep learning-based thoracic cavity segmentation for hyperpolarized 129Xe MRI
Suphachart Leewiwatwong1, Junlan Lu2, David Mummy3, Isabelle Dummer3,4, Kevin Yarnall5, Ziyi Wang1, and Bastiaan Driehuys1,2,3
1Biomedical Engineering, Duke University, Durham, NC, United States, 2Medical Physics, Duke University, Durham, NC, United States, 3Radiology, Duke University, Durham, NC, United States, 4Bioengineering, McGill University, Montréal, QC, Canada, 5Mechanical Engineering and Materials Science, Duke University, Durham, NC, United States
Quantifying hyperpolarized 129Xe MRI of pulmonary ventilation and gas exchange requires accurate segmentation of the thoracic cavity. This is typically done either manually or semi-automatically using an additional proton scan volume-matched to the gas image. These methods are prone to operator subjectivity, image artifacts, alignment/registration issues, and SNR. Here we demonstrate using a 3D convolutional neural network (CNN) to automatically and directly delineate the thoracic cavity from 129Xe MRI alone. This 3D-CNN uses a combination of Dice-Focal, perceptual loss, and training with template-based data augmentation to demonstrate thoracic cavity segmentation with a Dice score of 0.955 vs. expert readers.
|
||
 |
0392.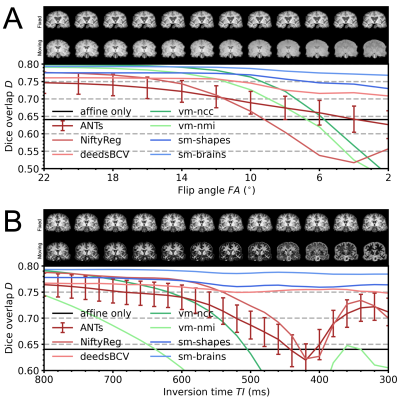 |
Learning-based non-linear registration robust to MRI-sequence contrast
Malte Hoffmann1,2, Benjamin Billot3, Juan Eugenio Iglesias1,2,3,4, Bruce Fischl1,2,4, and Adrian V Dalca1,2,4
1Department of Radiology, Harvard Medical School, Boston, MA, United States, 2Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 3Centre for Medical Image Computing, University College London, London, United Kingdom, 4Computer Science and Artificial Intelligence Laboratory, MIT, Cambridge, MA, United States
We introduce a novel strategy for learning deformable registration without acquired imaging data, producing networks robust to MRI contrast. While classical methods repeat an optimization for every new image pair, learning-based methods require retraining for accurate registration of unseen image types. To address these inefficiencies, we leverage a generative strategy for diverse synthetic label maps and images that enable training powerful networks that generalize to a broad spectrum of MRI contrasts. We demonstrate robust and accurate registration of arbitrary unseen MRI contrasts with a single network, thereby eliminating the need for retraining models.
|
|
 |
0393.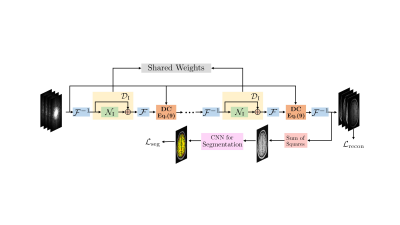 |
Image domain Deep-SLR for Joint Reconstruction-Segmentation of Parallel MRI
Aniket Pramanik1 and Mathews Jacob1
1Electrical and Computer Engineering, The University of Iowa, Iowa City, IA, United States
We present a novel framework for the joint reconstruction and segmentation of parallel MRI (PMRI) brain data. We introduce an image domain deep network for calibrationless recovery of undersampled PMRI data. It is deep-learning based generalization of local low-rank approaches for uncalibrated PMRI recovery including CLEAR. The image domain approach exploits additional annihilation relations compared to k-space based approaches and hence offers improved performance. To minimize segmentation errors resulting from undersampling artifacts, we combined the proposed scheme with a segmentation network and trained it end-to-end. It offers improved reconstruction with reduced blurring and sharper edges than independently trained reconstruction network.
|
|
 |
0394.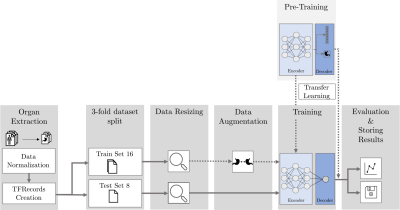 |
MRI-Based Response Prediction to Immunotherapy of Late-Stage Melanoma Patients Using Deep Learning
Annika Liebgott1,2, Louisa Fay1, Viet Chau Vu2, Bin Yang1, and Sergios Gatidis2
1Institute of Signal Processing and System Theory, University of Stuttgart, Stuttgart, Germany, 2Department of Radiology, University Hospital of Tuebingen, Tuebingen, Germany
The treatment of malignant melanoma with immunotherapy is a promising approach to treat advanced stages of the disease. However, the treatment can cause serious side effects and not every patient responds to it, which means crucial time may be wasted by an ineffective treatment. Assessment of the possible therapy response is hence an important research issue. The research presented in this study focuses on the investigation of the potential of medical imaging and machine learning to solve this task. To this end, we trained and compared different deep learning models on multi-modal PET/MR images to differentiate non-responsive from responsive patients.
|
|
 |
0395.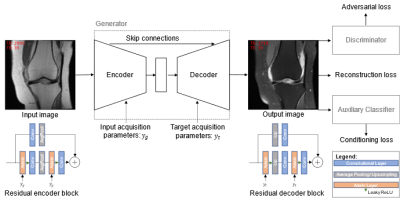 |
Fat-Saturated MR Image Synthesis with Acquisition Parameter-Conditioned Image-to-Image Generative Adversarial Network
Jonas Denck1,2,3, Jens Guehring3, Andreas Maier1, and Eva Rothgang2
1Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander University Erlangen-Nürnberg, Erlangen, Germany, 2Department of Industrial Engineering and Health, Technical University of Applied Sciences Amberg-Weiden, Weiden, Germany, 3Siemens Healthcare, Erlangen, Germany
We trained an image-to-image GAN that incorporates the sequence parameterizations in terms of the acquisition parameters repetition time and echo time into the image synthesis. We trained our model on the generation of synthetic fat-saturated MR knee images from non-fat-saturated MR knee images conditioned on the acquisition parameters, enabling us to synthesize MR images with varying image contrast. Our approach yields an NMSE of 0.11 and PSNR of 23.64, and surpasses the performance of a pix2pix [1] benchmark method. It can potentially be used to synthesize missing/additional MR contrasts and for customized data generation to support AI training.
|
|
0396.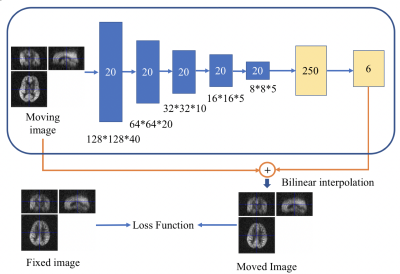 |
Image Registration of Perfusion MRI Using Deep Learning Networks
Zongpai Zhang1, Huiyuan Yang1, Yanchen Guo1, Lijun Yin1, David C. Alsop2, and Weiying Dai1
1State University of New York at Binghamton, Binghamton, NY, United States, 2Beth Israel Deaconess Medical Center & Harvard Medical School, Boston, MA, United States
Convolutional neural network (CNN) has demonstrated its accuracy and speed in image registration of structural MRI. We designed an affine registration network (ARN), based on CNN, to explore its feasibility on image registration of perfusion fMRI. The six affine parameters were learned from the ARN using both simulated and real perfusion fMRI data and the transformed images were generated by applying the transformation matrix derived from the affine parameters. The results demonstrated that our ARN markedly outperforms the iteration-based SPM algorithm both in simulated and real data. The current ARN is being extended for deformable fMRI image registration.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.