Bradley Fitzgerald1, Kausar Abbas2, Thomas M. Talavage1,3, and Joaquin Goni2
1Electrical & Computer Engineering, Purdue University, West Lafayette, IN, United States, 2Industrial Engineering, Purdue University, West Lafayette, IN, United States, 3Biomedical Engineering, University of Cincinnati, Cincinnati, OH, United States
1Electrical & Computer Engineering, Purdue University, West Lafayette, IN, United States, 2Industrial Engineering, Purdue University, West Lafayette, IN, United States, 3Biomedical Engineering, University of Cincinnati, Cincinnati, OH, United States
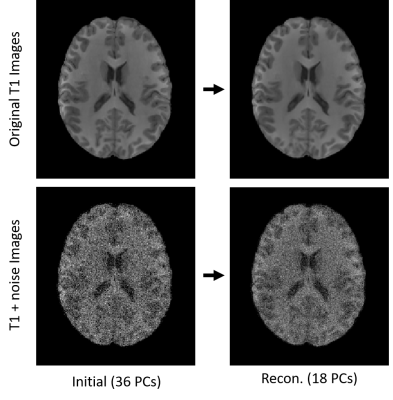
Differential identifiability of scan-rescan subject images can be maximized via principal component reconstruction of a T1 anatomical brain MRI dataset. This results in increased similarity between same subject repeated scans, as well as reducing apparent intersession noise in images.

Figure 1. Differential
identifiability and distance metrics for original T1 data and for T1 data with
added noise. (A) displays computed Idiff
values for original T1 images (black) and T1 images with added noise (red), reconstructed
with the full range of possible principle components (PCs). (B) displays Dself (blue) and
Dothers
(green) values for original T1 images and T1 images with added noise,
reconstructed with varying number of PCs.