Linfang Xiao1,2, Yilong Liu1,2, Zheyuan Yi1,2,3, Yujiao Zhao1,2, Peiheng Zeng1,2, Alex T.L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
This study presents a new MR
diagnostic paradigm where pathology detection is performed directly from
extremely sparse k-space data and provides an extremely rapid and potentially
powerful tool for automatic pathology screening.
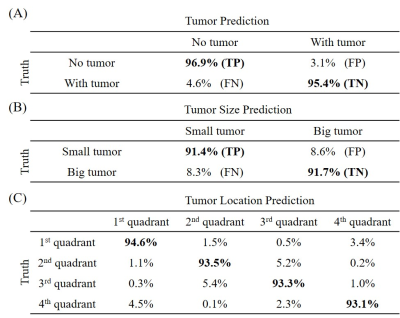
Figure 3. ANN prediction results from a single-shot spiral trajectory with 251 points for (A) tumor
detection, (B) tumor size
classification, and (C) tumor
location classification.

Figure 2. The
proposed ANN model structure for tumor detection, and size/location
classification. The model input is a vector concatenated by the real and
imaginary part of the complex k-space data, outputting three categories of
probabilities. Three tasks share the first two fully connected layers (FC), and
each FC is followed by a batch normalization layer, a rectified linear unit
(ReLU) layer and a dropout layer (50%). Each task has its own fully connected
layer and softmax layer for classification. Noted that tumor size/location is
only classified in presence of tumors.