Digital Poster
Modeling I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1415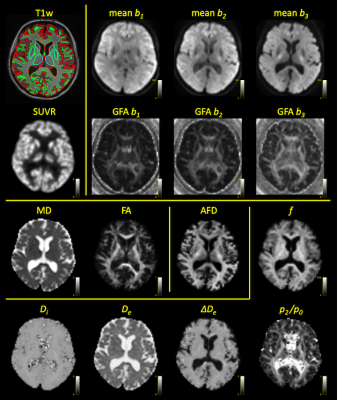 |
39 | Correlating neurite density and synaptic density in the human brain in vivo with diffusion-weighted PET-MR
Daan Christiaens1,2, Thomas Vande Casteele2,3,4, Maarten Laroy2,4, Margot Van Cauwenberge2,3,4, Jan Van den Stock3,4, Filip Boeckaert3,4, Stefan Sunaert2,5,6, Mathieu Vandenbulcke3,4, Frederik Maes1,2, and Louise Emsell2,3,4,5
1Department of Electrical Engineering, ESAT/PSI, KU Leuven, Leuven, Belgium, 2Medical Imaging Research Center, University Hospitals Leuven, Leuven, Belgium, 3Geriatric Psychiatry, UPC KU Leuven, Leuven, Belgium, 4Leuven Brain Institute, Department of Neurosciences, Neuropsychiatry, KU Leuven, Leuven, Belgium, 5Department of Imaging & Pathology, Translational MRI, KU Leuven, Leuven, Belgium, 6Radiology, University Hospitals Leuven, Leuven, Belgium
11C-UCB-J PET offers a unique imaging modality to map synaptic density in the human brain in vivo with high specificity. Here, we investigate its correlation with several diffusion MRI metrics and microstructure model parameters in diffusion-weighted PET-MR. We report moderate negative correlation of 11C-UCB-J uptake with measures of anisotropy, consistent with a hypothesis that higher synaptic density is associated with a more disorganised neurite configuration. We also find weak positive correlation to the intra-axonal signal fraction in cortical grey matter. As such, 11C-UCB-J PET-MR can further the interpretation and in vivo validation of more advanced microstructure models of grey matter.
|
||
1416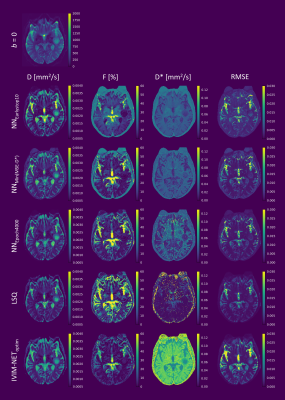 |
40 | The impact of learning rate, network size, and training time on unsupervised deep learning for intravoxel incoherent motion (IVIM) model fitting
Misha Pieter Thijs Kaandorp1,2, Frank Zijlstra1,2, João P. de Almeida Martins1,2, Christian Federau3,4, and Peter T. While1,2
1Department of Radiology and Nuclear Medicine, St. Olav’s University Hospital, Trondheim, Norway, 2Department of Circulation and Medical Imaging, NTNU – Norwegian University of Science and Technology, Trondheim, Norway, 3Institute for Biomedical Engineering, University and ETH Zürich, Zurich, Switzerland, 4AI Medical, Zürich, Switzerland
We demonstrate that a high learning rate, small network size, and early stopping in unsupervised deep learning for IVIM model fitting can result in sub-optimal solutions and correlated parameters. In simulations, we show that prolonging training beyond early stopping resolves these correlations and reduces parameter error, providing an alternative to exhaustive hyperparameter optimization. However, extensive training results in increased noise sensitivity, tending towards the behavior of least squares fitting. In in-vivo data from glioma patients, fitting residuals were almost identical between approaches, whereas pseudo-diffusion maps varied considerably, demonstrating the difficulty of fitting D* in these regions.
|
||
1417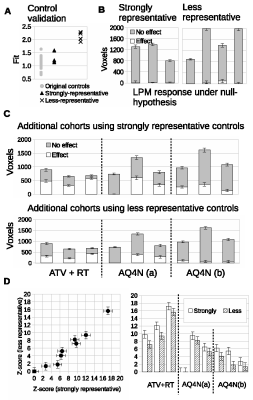 |
41 | Improved Modelling of ADC Distributions makes Therapeutic co-clinical Trials possible with very small animal numbers
Paul Tar1, N.A. Thacker2, M. Babur2, G. Lipowska-Bhalla2, S. Cheung2, R. Little2, K.J. Williams2, and J.P.B. O'Connor2
1Cancer, University of Manchester, Manchester, United Kingdom, 2University of Manchester, Manchester, United Kingdom
In oncology, preclinical experiments using MRI often evaluate spatially complex and heterogeneous tumor micro-environments which have non-Gaussian data and small sample sizes, with cohorts typically of 10 animals or less. As a consequence, conventional use of t-tests that evaluate distribution parameters such as means and percentiles can be ineffective. Further, the cohort-level nature of such analyses also limits investigations to groups of tumors rather than identifying individually responding tumors. In contrast, Linear Poisson Modelling (LPM) enables quantitative analysis of complex data, can operate in small data domains and can also provide per-tumor assessments 2.ideal for co-clinical trials.
|
||
1418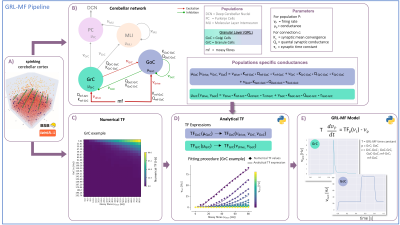 |
42 | GRL-MF: The first lego-brick of a cerebellar Mean-Field Model for BOLD signal simulations
Roberta Maria Lorenzi1, Alice Geminiani1, Claudia AM Gandini Wheeler-Kingshott1,2,3, Fulvia Palesi1, Claudia Casellato1, and Egidio D'Angelo1,3
1Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 2NMR Research Unit, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, UCL, London, United Kingdom, 3Brain Connectivity Center, IRCCS Mondino Foundation, Pavia, Italy
Following MRI recordings, the connectome can be reconstructed and BOLD-signals simulated extracting relevant parameters on brain organization and dynamics. This requires mathematical models of neural activity in specific brain regions. However, models of the cerebellar circuit are still missing. We present here a biologically-driven mean-field (MF) model summarising the main statistical moments of cerebellar granular layer activity as the first “lego-brick” toward full cerebellar network reconstruction. Once integrated into brain simulators, like Dynamic Causal Modelling and The Virtual Brain, the cerebellar MF models will improve the investigation of neuronal functions at the origin of hemodynamic responses captured by BOLD fMRI.
|
||
1419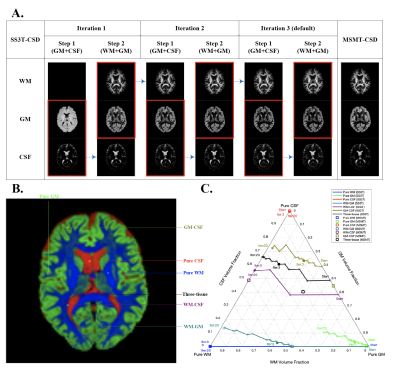 |
43 | Evaluation of SS3T-CSD for the analysis of diffusion MRI data - a simulation study
Jibin Tang1, Maximilian Pietsch2,3, and Jacques-Donald Tournier2,4
1Department of Medical Engineering and Physics, School of Biomedical Engineering and Imaging Sciences, Faculty of Life Sciences and Medicine, King's College London, London, United Kingdom, 2Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 3MRC Centre for Neurodevelopmental Disorders, King's College London, London, United Kingdom, 4Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Single-shell three-tissue constrained spherical deconvolution (SS3T-CSD) has been proposed to decompose the dMRI signal into three different tissue types from single-shell data. Here, we evaluate the SS3T-CSD method over a range of different situations using real data and simulations, varying parameters including SNR, partial volume effect, b-value, number of iterations, and tissue response functions. We evaluate their effect on results by comparing with the ground truth and the results estimated from MSMT-CSD. Our results indicate that while SS3T-CSD performs well, it is sensitive to some of these factors and shows some deficiencies in certain situations.
|
||
1420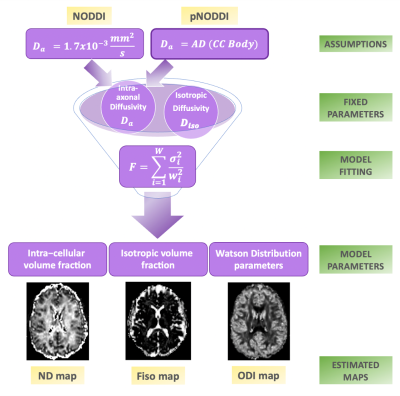 |
44 | A personalized-NODDI (pNODDI) pipeline increases sensitivity to microstructural alterations in temporal lobe epilepsy
Elena Grosso1, Claudia AM Gandini Wheeler-Kingshott1,2,3, Egidio D'Angelo1,3, Paolo Vitali4, and Fulvia Palesi1
1Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 2NMR Research Unit, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, UCL, London, United Kingdom, 3Brain Connectivity Centre Research Unit, IRCCS Mondino Foundation, Pavia, Italy, 4Radiology, IRCCS Policlinico San Donato, San Donato Milanese, Italy
Several mesoscopic multi-compartment diffusion MRI models have been developed to describe the complexity of water molecules diffusion in the brain by making assumptions. One frequently used model is Neurite Orientation Dispersion and Density Imaging (NODDI). Here, the aim is to propose a personalized-NODDI (pNODDI) by making NODDI assumptions subject-specific. Our findings show that NODDI assumptions impact on output metrics depending on brain region and that these metrics are able to discriminate between healthy and temporal lobe epilepsy subjects. pNODDI provides a mean to personalize a successful and clinically feasible model as NODDI, increasing metrics sensitivity to pathological alterations.
|
||
1421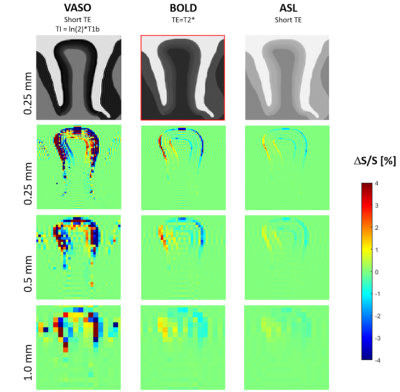 |
45 | Effect of brain tissue deformation on functional MRI signal variations assessed using biomechanical simulations Video Permission Withheld
Mahsa Zoraghi1, Nico Scherf2,3, Carsten Jaeger1, Ingolf Sack4, Sebastian Hirsch5,6, Stefan Hetzer5,6, and Nikolaus Weiskopf1,7
1Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Methods and Development Group Neural Data Science and Statistical Computing, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 3Institute for Medical Informatics and Biometry, Carl Gustav Carus Faculty of Medicine, TU Dresden, Dresden, Germany, 4Department of Radiology, Charité – Universitätsmedizin Berlin, Berlin, Germany, Berlin, Germany, 5Berlin Center for Advanced Neuroimaging, Charité – Universitätsmedizin Berlin, Berlin, Germany, 6Berlin Center for Computational Neuroscience, Berlin, Germany, 7Faculty of Physics and Earth Sciences, Felix Bloch Institute for Solid State Physics, Leipzig University, Leipzig, Germany
Recent studies on the brain suggest a link between brain function and biomechanics of brain. In this study we develop a novel simulation framework to investigate blood flow-induced deformation caused by increased neural activity in brain tissue considering its mechanical properties. We further investigate its impact in simulated fMRI experiments. Our results demonstrate that the displacement in gyrus induced by local volume and stiffness change might not be directly resolved due to limited fMRI resolution, but can lead to artifacts when interpreting measurements at layer-level resolution. Our findings may help to systematically analyze potential resulting artefacts in high resolution fMRI.
|
||
1422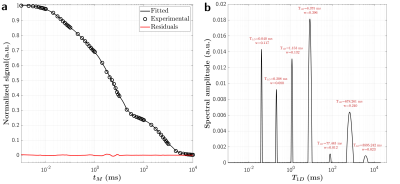 |
46 | Quantitative T1D assessment in lipid membranes: Jeener-Broekaert NMR vs. ihMT MRI
Andreea Hertanu1,2, Lucas Soustelle1,2, Ludovic de Rochefort1,2, Axelle Grélard3, Antoine Loquet3, Erick J. Dufourc3, Gopal Varma4, David C. Alsop4, Guillaume Duhamel1,2, and Olivier M. Girard1,2
1Aix Marseille Univ, CNRS, CRMBM, Marseille, France, 2APHM, Hôpital Universitaire Timone, CEMEREM, Marseille, France, 3CBMN UMR 5248, CNRS, University of Bordeaux, Bordeaux INP, Pessac, France, 4Division of MR Research, Radiology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
The dipolar order relaxation time (T1D) is a probe of membrane dynamics and microstructure and could serve to further understand the relationship between the myelin membrane integrity and its biological function. In this work, the ability of quantitative inhomogeneous Magnetization Transfer (qihMT) to estimate the several T1D components of a synthetic lipid membrane system, a surrogate for the myelin membrane, was evaluated by comparison with the gold standard method for T1D quantification, the NMR Jeener-Broekaert (JB) sequence.
|
||
1423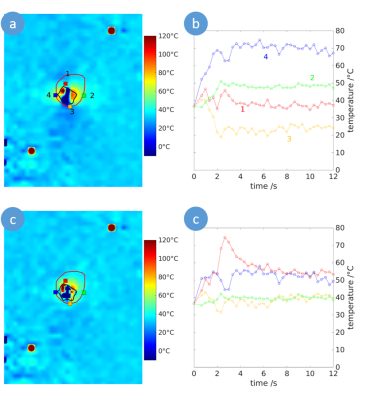 |
47 | Susceptibility corrected Proton Resonance Frequency Shift based MR Thermometry of hepatic Microwave Ablation in an in-vivo swine model
Marcel Gutberlet1,2, Enrico Pannicke2,3, Inga Bruesch4, Regina Rumpel4, Eva-Maria Wittauer4, Florian W. R. Vondran5, Frank Wacker1,2, and Bennet Hensen1,2
1Institute of Diagnostic and Interventional Radiology, Medical School Hannover, Hannover, Germany, 2STIMULATE-Solution Centre for Image Guided Local Therapies, Magdeburg, Germany, 3Department Biomedical Magnetic Resonance, Otto-von-Guericke University, Magdeburg, Germany, 4Institute for Laboratory Animal Science and Central Animal Facility, Medical School Hannover, Hannover, Germany, 5Clinic for General, Abdominal and Transplant Surgery, Medical School Hannover, Hannover, Germany
In microwave ablation (MWA), heat induced susceptibility changes impair the assessment of the ablation zones using proton resonance frequency shift based magnetic resonance (MR) thermometry. In this work, these heat related field changes were modelled to improve the accuracy of MR thermometry to monitor microwave ablation. In a study of hepatic MWA in an in-vivo swine model, the proposed method provided increased accuracy to assess the ablation zone compared to uncorrected MR thermometry.
|
||
1424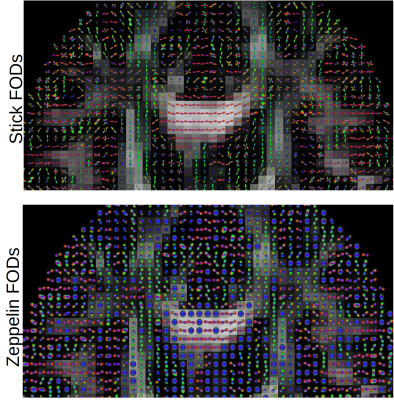 |
48 | Voxelwise estimation of multiple response functions and fiber orientation distribution functions in diffusion MRI
Samuel St-Jean1,2, Alexander Leemans3, Filip Szczepankiewicz1, Christian Beaulieu2, and Markus Nilsson1
1Clinical Sciences Lund, Lund University, Lund, Sweden, 2University of Alberta, Edmonton, AB, Canada, 3Image Sciences Institute, UMC Utrecht, Utrecht, Netherlands
We present a framework to compute a response function for each voxel using a fingerprinting approach and a per voxel, per compartment estimation of the fiber orientation distribution (FOD) function for diffusion MRI. The method first matches the powder-averaged signal to a database of candidate signals and estimates a unique FOD from this matched signal for each compartment. This is done by turning the candidate signals into continuous Legendre polynomials, leading to a set of linear equations to estimate coefficients of the FODs, without requiring a prior segmentation or averaging multiple response functions.
|
||
1425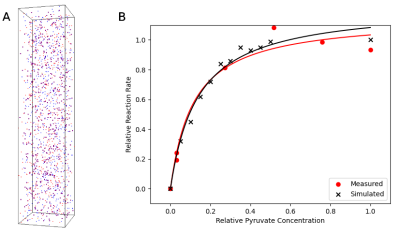 |
49 | Novel particle-based spatial stochastic Bloch simulation applied to LDH-mediated pyruvate conversion
Dylan Archer Dingwell1,2 and Charles H Cunningham1,2
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Physical Sciences, Sunnybrook Research Institute, Toronto, ON, Canada
In order to model signal mechanisms relevant to HP 13C MRI of lactate, we developed a novel particle-based MR model which extends the Brownian dynamics simulator Smoldyn. The model performs concurrent calculation of a forward solution of the Bloch equations for simulated particles using quaternion rotations. Reaction kinetics of LDH-mediated conversion of pyruvate to lactate were simulated and compared to a benchtop experiment (LDH activity assay). Modelling of particle compartmentalization and motion were also tested in simulated structures.
|
||
1426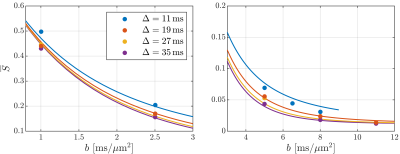 |
50 | Time dependence at ultra-high diffusion weighting reveals fast compartmental exchange in rat cortex in vivo
Jonas Lynge Olesen1,2, Andrada Ianus3, Noam Shemesh3, and Sune Nørhøj Jespersen1,2
1Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Aarhus University, Aarhus, Denmark, 2Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark, 3Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal
The potential effect of compartmental water exchange and somas on the diffusion MRI signal in grey matter tissue are currently open questions with considerable implications for biophysical modelling of grey matter at (pre-)clinical diffusion time scales. Recent studies have begun to tackle these questions and found evidence that compartmental water exchange dominates the diffusion time dependence. Here, we extend the application of the Standard Model with exchange between the neurites and extracellular water (SMEX) to in vivo rat brain and again find evidence for exchange-dominated time dependence with very short neurite residence time on the order of 2 ms.
|
||
1427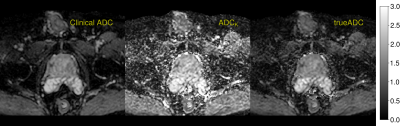 |
51 | A novel clinical prostate diffusion MRI approach for combined measurement of ADC and non-monoexponential diffusion
Stefan Kuczera1, Fredrik Langkilde1, and Stephan E. Maier1,2
1Institute of Clinical Sciences, Sahlgrenska Academy, Gothenburg University, Gothenborg, Sweden, 2Department of Radiology, Brigham Women’s Hospital, Harvard Medical School, Boston, MA, United States
A novel clinical protocol for prostate diffusion imaging is presented. The commonly applied strategy of averaging MR signal at a small number of b-values is replaced by the acquisition of a whole range of b-values in conjunction with a model fitting post precessing step. With no increase in acquisition time, our results show that the same image quality can be achieved. As a main benefit, modelling is made more consistent and largely b-value independent for both the simple ADC model, as well as for more complex signal representations such as the kurtosis model.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.