Digital Poster
Cardiac Anatomy & Tissue Characterization VI
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1836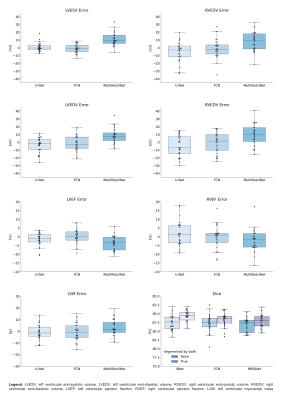 |
16 | Multilevel Comparison of Neural Networks for Ventricular Function Quantification in CMR accelerated by Compressed Sensing
Thomas Hadler1, Clemens Ammann1, Jan Gröschel1,2, and Jeanette Schulz-Menger1,2,3
1Charité - Universitätsmedizin Berlin, Working Group on CMR, Experimental and Clinical Research Center, a joint cooperation between the Max-Delbrück-Center for Molecular Medicine and the Charité – Universitätsmedizin Berlin, Berlin, Germany, 2DZHK (German Centre for Cardiovascular Research), partner site Berlin, Berlin, Germany, 3Department of Cardiology and Nephrology, Helios Hospital Berlin-Buch, Berlin, Germany
Three popular convolutional neural networks were trained on short-axis cine MR images of the heart acquired by a prototype 2-shot 2D Compressed Sensing sequence. Network performance was evaluated on the level of clinical results and segmentation quality. Analysis revealed high correlation for quantitative results between all networks and a human expert. Automatic segmentation of the right ventricle is significantly more difficult than the left ventricle and shows more outliers. Segmentation decision errors concentrate in basal and apical slices, with the largest millilitre differences in the basal slices. Fast acquisition and automated image analysis promise high efficiency in CMR.
|
||
1837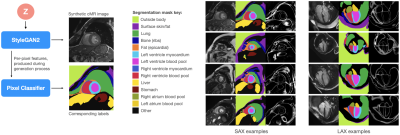 |
17 | Semantic CMR Synthesis: Generating Coherent Short- and Long-Axis Images with Corresponding Multi-Class Anatomical Labels
Thomas Joyce1, Nico Schulthess1, Gloria Wolkerstorfer1, Stefano Buoso1, and Sebastian Kozerke1
1University and ETH Zurich, Zurich, Switzerland
We propose to use a combination of the StyleGAN2, ADA and DatasetGAN methods to produce synthetic short- and long-axis view cardiac magnetic resonance (CMR) images accompanied with corresponding 11-class tissue masks. The image generator networks are trained on datasets of approximately 1850 and 5000 unlabelled images, for short- and long-axis images respectively. The segmentation networks are trained on only 30 manually annotated synthetic images in total. We further demonstrate a proof-of-concept method for generating coherent long- and short-axis images of the same synthetic patient.
|
||
1838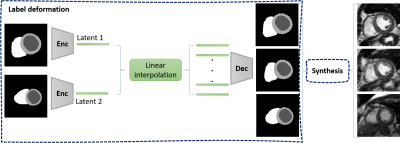 |
18 | Intra- and intersubject synthesis of cardiac MR images using a VAE and GAN
Sina Amirrajab1, Cristian Lorenz2, Jürgen Weese2, Josien Pluim1, and Marcel Breeuwer1,3
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands We propose a method for synthesizing cardiac MR images with plausible heart shape and realistic appearance. It breaks down the synthesis into labels deformation and label-to-image translation. The former is achieved via latent space interpolation in a VAE model, while the latter is accomplished via a conditional GAN model. We synthesize 32 short-axis slices within each subject (intrasubject), as well as eight intermediary generated subjects between two dissimilar real subjects (intersubject) that have different anatomies. Such method could provide a solution to enrich a database and to pave the way for the development of generalizable DL based algorithms. |
||
1839 |
19 | Accelerating multi-contrast multidimensional motion-corrected non-Cartesian iterative reconstructions with the Toeplitz approach
Andrew Phair1, Gastao Cruz1, René Botnar1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Iterative reconstructions for multi-contrast multidimensional non-Cartesian MRI can require considerable computation time, with a large fraction dedicated to non-uniform fast Fourier transforms (NUFFTs). The Toeplitz approach has been previously shown to allow NUFFT operations to be replaced by computationally efficient fast Fourier transforms (FFTs) without loss of accuracy. Herein, we combine this approach with a 3D multi-contrast low-rank cardiac-motion-corrected radial reconstruction with low-rank patch-based regularisation to achieve a ~13.3-fold computational speed-up.
|
||
1840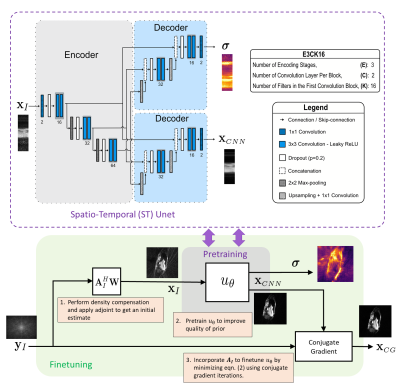 |
20 | Data-Efficient Uncertainty Quantification for Radial Cardiac Cine MR Image Reconstruction
Sherine Brahma1, Tobias Schäffter1,2,3, Christoph Kolbitsch1,3, and Andreas Kofler1
1Physikalisch-Technische Bundesanstalt, Braunschweig and Berlin, Germany, 2Department of Biomedical Engineering, Technical University of Berlin, Berlin, Germany, 3School of Imaging Sciences and Biomedical Engineering, King’s College London, London, United Kingdom
Due to the black-box nature of Deep Learning (DL) algorithms, uncertainty quantification (UQ) is a promising approach to assess their risk in medical applications. However, UQ is challenging in imaging techniques like non-Cartesian multi-coil CINE MRI because the data is high-dimensional, and the acquisition process is computationally demanding. In this work, i) we propose to utilize spatio-temporal (ST) networks, demonstrating efficient UQ in a high-dimensional setting, and ii) we show a reduction in uncertainty by adopting the forward model of radial multi-coil 2D cine MR in the reconstruction process. UQ is performed using MCMC dropout with additional aleatoric loss terms.
|
||
1841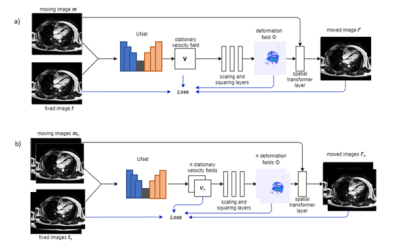 |
21 | Diffeomorphic Image Registration for CINE Cardio MR images using deep learning
Daniel Amsel1, Seung Su Yoon1,2, Jens Wetzl2, and Andreas Maier1
1Friedrich-Alexander University Erlangen-Nuernberg, Erlangen, Germany, 2Siemens-Healthineers, Erlangen, Germany Diagnostic applications often require the estimation of organ motion. Image registration enables motion estimation by computing deformation fields for an image pair. In this work, voxelmorph, a framework for deep learning-based diffeomorphic image registration is used to register CINE cardiac MR images in four-chamber view. Additionally, the framework is extended to a one-to-many registration to also utilize temporal information within a time-resolved MR scan. Registration performance as well as the performance of a valve tracking application using this approach are evaluated. The results are comparable to a state-of-the-art registration method, while noticeably reducing the computation time. |
||
1842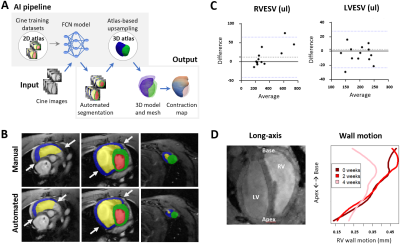 |
22 | Automated cardiac bi-ventricular segmentation and motion analysis in a monocrotaline rat model of pulmonary hypertension Video Permission Withheld
Marili Niglas1, Nicoleta Baxan2, Ali Ashek1, Lin Zhao1, Jinming Duan3, Declan O'Regan4, Timothy JW Dawes1,4, Wenjia Bai5, and Lan Zhao1
1National Heart and Lung Institute, Imperial College London, London, United Kingdom, 2Biological Imaging Centre, Imperial College London, London, United Kingdom, 3School of Computer Science, University of Birmingham, London, United Kingdom, 4MRC London Institute of Medical Sciences, Imperial College London, London, United Kingdom, 5Department of Brain Sciences, Imperial College London, London, United Kingdom
Multiple cardiac AI-based image processing pipelines exist for clinical application, yet the equivalent is lacking for rodents. We utilized a fully convolutional network combined with 3D-atlas registration to auto-segment cine images from pulmonary hypertension (PH) rats and produce 3D contraction maps. The auto-segmentations were equivalent to manual (Dice overall >0.7). The volumetric parameters did not differ between methods, except a minor underestimation for RVESV in PH rat (8.2%). 3D contraction maps indicated moderately increased basal wall motion at early (adaptive) stage followed by a 36% reduction at later (maladaptive) stage of PH. This regional motion remodelling correlates with PAH patients.
|
||
1843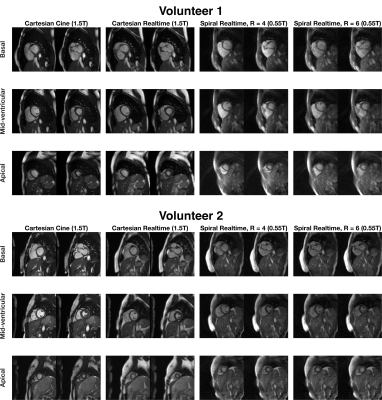 |
23 | Real-time Cardiac MRI at 0.55T using through-time spiral GRAPPA
Alexander Fyrdahl1 and Nicole Seiberlich1
1Radiology, University of Michigan, Ann Arbor, MI, United States
Through-time non-Cartesian GRAPPA can be used to enable real-time imaging for evaluation of left-ventricular function. However, it requires arrays with many receiver coils for the parallel imaging reconstruction and gradient systems capable of playing out the specified non-Cartesian trajectories. In this work, we explore the use of non-Cartesian GRAPPA for real-time cardiac imaging on a 0.55T system with lower performance gradients, reduced receiver coils compared to conventional MRI systems.
|
||
1844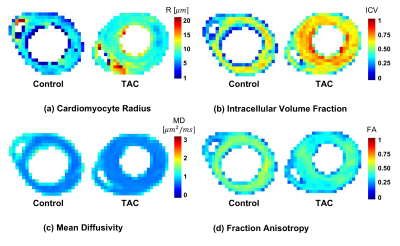 |
24 | Quantifying Cardiac Hypertrophy using Biophysical Models of Diffusion MRI
Mohsen Farzi1, Darryl McClymont2, Hannah Whittington2, Marie-Christine Zdora3, Leah Khazin1, Craig A. Lygate2, Christoph Rau3, Erica Dall’Armellina1, Irvin Teh1, and Jürgen E. Schneider1
1Biomedical Imaging Science Department, University of Leeds, Leeds, United Kingdom, 2Cardiovascular Medicine, University of Oxford, Oxford, United Kingdom, 3Diamond Light Source Ltd., Oxfordshire, United Kingdom
Biophysical modelling is a valuable technique for interrogating cardiac microstructure. We utilise a two-compartment model where intra- and extra-cellular space were represented by impermeable cylinders and an oblate diffusion tensor, respectively. Data was acquired ex vivo using a bespoke diffusion scheme with five diffusion times, six b-values and ten directions. Cardiomyocyte radius (r = 7.6 versus 8.9 μm) and volume fraction (ICV = 0.56 versus 0.67) were sensitive markers of cardiomyopathy validated in one healthy and one disease heart with transverse aortic constriction (TAC).
|
||
1845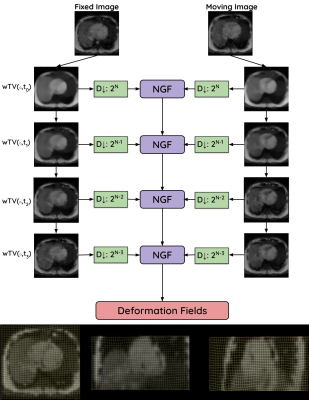 |
25 | 3D Multiscale Weighted Total Variation Registration for MR Image-Guided Catheter Interventions
Jaykumar Patel1,2, Calder Sheagren1,2, Saqeeb Hassan2, Fatemeh Rastegar Jouybari1,3, Christopher Macgowan1,3, and Graham Wright1,2
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Schulich Heart Centre, Sunnybrook Research Institute, Toronto, ON, Canada, 3The Hospital for Sick Children, Toronto, ON, Canada
During MR-guided cardiac catheter intervention, the catheter may be misaligned due to the respiratory motion of the heart. To improve the myocardial border alignment, GPU-based edge-preserving image registration is developed to accurately align highly undersampled 3D cone-trajectory image-based navigators for motion characterization. The edge-preserving registration technique improved myocardial border alignment across differing spatial resolutions and acceleration factors in-silico.
|
||
1846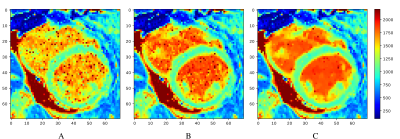 |
26 | Combining machine learning and mathematical modeling in estimation of T1 relaxation time
Kateřina Škardová1, Radek Galabov1,2, Kateřina Fricková1, Tomáš Pevný3, Jaroslav Tintěra2, Tomáš Oberhuber1, and Radomír Chabiniok1,4
1Department of Mathematics, FNSPE CTU in Prague, Prague, Czech Republic, 2Department of Radiology, Institute for clinical and experimental medicine, Prague, Czech Republic, 3Artificial Intelligence Center, CTU in Prague, Prague, Czech Republic, 4Department of Pediatrics, UT Southwestern Medical Center, Dallas, TX, United States A method for estimating tissue parameters using cardiovascular MRI and biophysical model by combining neural network (NN) and numerical optimization (NO) is illustrated on estimating $$$T_1$$$ relaxation time from MOLLI. Compared to the estimation obtained from MOLLI by the scanner, the proposed method provided $$$T_1$$$ closer to turbo spin-echo pseudo-ground in 7 out of 8 phantoms and higher or comparable myocardial and blood $$$T_1$$$ in 6 out of 7 patiens’ datasets. Including the NN-based initial guess accelerated the subsequent NO. NO initialized by NN, trained using simulated data, showed the potential to increase the efficiency and robustness of parameter estimation. |
||
1847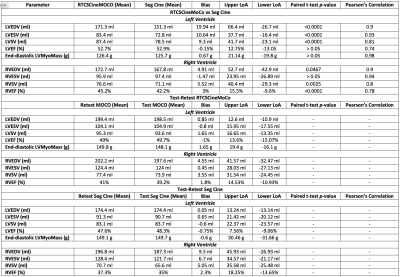 |
27 | Biventricular function using a novel real-time free-breathing compressed sensing cine sequence with R-R normalization and motion correction
Ahmed S Negm1, Jianing Pang2, Kiaran Mcgee1, Holly Iverson1, Maria Halverson1, Rachel Davids2, Prabhakar Rajiah1, Alexander Bratt1, Eric Williamson1, Phillip Young1, Thomas Foley1, Tim Leiner1, Christopher Francois1, Xiaoming Bi2, and Jeremy Collins3
1Radiology, Mayo Clinic, Rochester, MN, United States, 2Siemens Healthineers, Chicago, IL, United States, 3Mayo Clinic, Rochester, MN, United States CMR is the gold standard for assessing biventricular size and function. bSSFP Seg Cine is a standard cine method, but requires breath-holding which prolongs table time and limits utility in dyspneic patients. A novel real-time technique RTCSCineMoCo combines free-breathing acquisition with compressed sensing and motion correction. In 100 subjects RTCSCineMoCo generated similar results to Seg Cine for LVEF, LV end-diastolic mass, and RVESV with agreement including no difference at Bland-Altman analysis for all LV and RV measures. The shorter table time for RTCSCineMoCo and ability to robustly image dyspneic patients are important enhancements for clinical practice. |
||
1848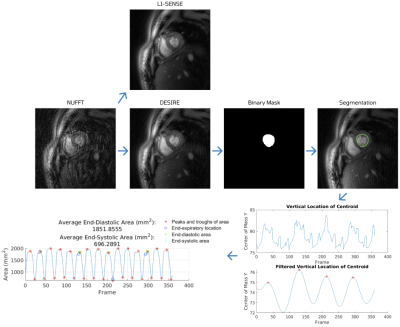 |
28 | Deep learning-based Automatic Analysis for Free-breathing High-resolution Spiral Real-time Cardiac Cine Imaging at 3T
Marina Awad1, Junyu Wang1, Xue Feng1, Ruixi Zhou1, and Michael Salerno1,2,3,4
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Medicine, University of Virginia, Charlottesville, VA, United States, 3Radiology, University of Virginia, Charlottesville, VA, United States, 4Medicine, Stanford University, Stanford, CA, United States Cardiac real-time cine imaging is useful for patients who cannot hold their breath or have irregular heart rhythms. Free breathing high-resolution real-time cardiac cine images are acquired efficiently using spiral acquisitions, and rapidly reconstructed using our DESIRE framework. However, quantifying the EF from free-breathing real-time imaging is limited by the lack of an EKG signal to define the cardiac cycle, and through-plane cardiac motion resulting from free-breathing. We developed a DL-based segmentation technique to determine the end-expiratory phase and determine end-systole and end-diastole on a slice by slice basis to accurately quantify EF from spiral real-time cardiac cine imaging. |
||
1849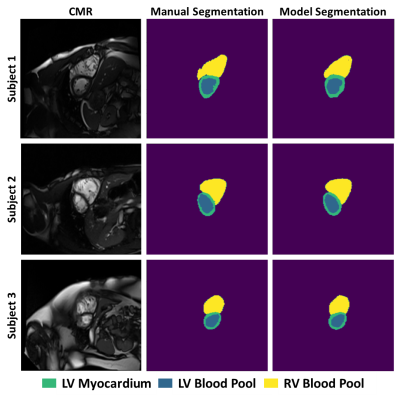 |
29 | Feasibility of machine learning for cardiovascular function analysis in patients with repaired tetralogy of Fallot
Elizabeth Walker Thompson1, Abhijit Bhattaru1, Phuong Vu1, Elizabeth Donnelly2, Elizabeth Goldmuntz2, Mark Fogel2, and Walter Witschey1
1University of Pennsylvania, Philadelphia, PA, United States, 2Children's Hospital of Philadelphia, Philadelphia, PA, United States
Tetralogy of Fallot (ToF) is a congenital heart disease that is typically repaired with surgery early in life, but right ventricular remodeling results in adverse events for many patients. This preliminary analysis of 8 patients investigated the feasibility of training a convolutional neural network to segment the right and left ventricles from 2-dimensional cardiovascular magnetic resonance images, resulting in Dice scores ranging from 0.73-0.91 for the left ventricular blood pool, left ventricular myocardium, and right ventricular blood pool. Machine learning shows promise to enable large-scale longitudinal studies of ToF.
|
||
1850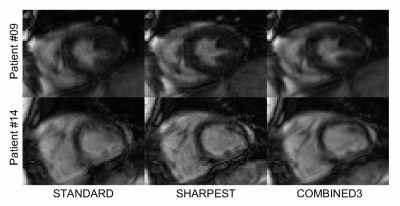 |
30 | Retrospective motion correction in multiple average, free-breathing cardiac cine imaging
Alexander Paul Neofytou1, Radhouene Neji1,2, James Wong3, Anastasia Fotaki1, Joana Ferreira1, Carl Evans1, Filippo Bosio1, Nabila Mughal1, Reza Razavi1, Kuberan Pushparajah1, and Sébastien Roujol1
1School of Biomedical Engineering and Imaging Sciences, Faculty of Life Sciences and Medicine, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom, 3Department of Paediatric Cardiology, Evelina London Children’s Hospital, London, United Kingdom
A novel reconstruction technique based on iterative rejection of segmented k-space was developed for retrospective correction of respiratory motion in multiple average, free-breathing cine images. In comparison to standard signal averaging reconstruction, it provides higher sharpness, SNR, CNR, image quality, and rate of diagnostic quality images.
|
||
1851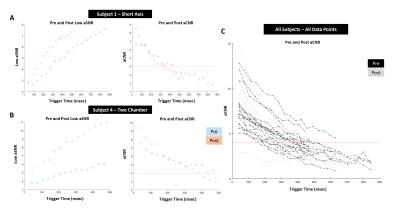 |
31 | Impact of Gadolinium injection to the variable Flip Angle scheme of Tag CMR including SENC and SPAMM
Vivian S. Nguyen1, Jacob P. Goes1, Donovan Gorre2, Hui Wang3,4, Marcella K. Vaicik5, Amit R. Patel6, and Keigo Kawaji1,6
1Biomedical Engineering, Illinois Institute of Technology, Chicago, IL, United States, 2Radiology, University of Chicago Medical Center, Chicago, IL, United States, 3Philips, Gainesville, FL, United States, 4Department of Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 5Illinois Institute of Technology, Chicago, IL, United States, 6Medicine - Cardiology, University of Chicago Medical Center, Chicago, IL, United States
SENC and SPAMM tagging Cardiac Magnetic Resonance (CMR) are specifically optimized for acquisition before contrast injection and tuned to the end-systolic phase. While SPAMM tagging is a non-quantitative approach that indirectly derives functional motion from the displacement of the saturated tags in the myocardium, SENC directly computes myocardial tissue compression (strain) in percentage units. Furthermore, SENC quantitation pipeline bypasses the signal intensity map that exhibits the decay of the embedded strain measurements. In this study, we examine both SPAMM and SENC post-Gd measurements to specifically indicate the effect of the variable RF pulse train tuned to target tissue T1.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.