Digital Poster
Processing & Analysis: Software Tools & Visualization II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
2769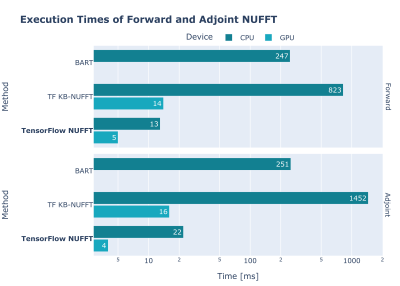 |
37 | TensorFlow MRI: A Library for Modern Computational MRI on Heterogenous Systems
Javier Montalt-Tordera1, Jennifer Steeden1, and Vivek Muthurangu1
1University College London, London, United Kingdom
We present TensorFlow MRI, a new open-source library of TensorFlow operators for MR image reconstruction and processing. Its goal is to enable fast prototyping of modern MRI applications within a single computing framework. It is intended for researchers working in MR image reconstruction and/or processing, especially those interested in ML applications. The library is primarily Python-based and is easy to use, understand and extend. It has a single-command installation procedure and extensive documentation. Thanks to the use of a TensorFlow backend, it has excellent performance and runs on heterogenous and distributed systems.
|
||
2770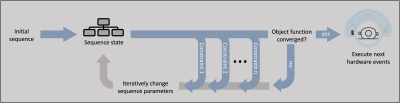 |
38 | Constraint-Based Sequence Optimization in a Scanner-Independent MRI Framework
Daniel Christopher Hoinkiss1, Simon Konstandin1,2, and Matthias Günther1,2,3
1Fraunhofer Institute for Digital Medicine MEVIS, Bremen, Germany, 2mediri GmbH, Heidelberg, Germany, 3University of Bremen, Bremen, Germany
We demonstrate a constraint-based approach of MRI sequence development in the vendor-independent MRI pulse sequence development framework gammaSTAR and demonstrate this concept in arterial spin labeling by optimizing the timings of the background suppression pulses to minimize the signal contribution of chosen T1 values in the human brain. This concept can raise MRI sequence development to a new abstraction level by, instead of providing exact timings and parameter values, defining physical constraints and conditions to be satisfied by the MRI sequence during an automated sequence generation process.
|
||
2771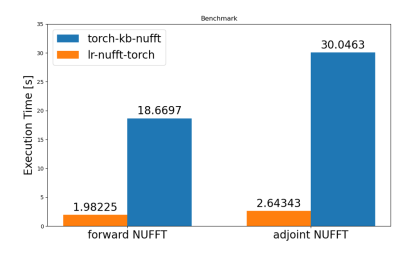 |
39 | Efficient Non-Uniform Fourier Transform with embedded low-rank projection for fast model-based MRI reconstruction
Matteo Cencini1,2, Luca Peretti2,3, and Michela Tosetti1,2
1IRCCS Stella Maris, Pisa, Italy, 2Imago7 Foundation, Pisa, Italy, 3Università di Pisa, Pisa, Italy
Modern approaches to iterative imaging, such as model-based reconstruction, requires efficient implementations of Non-Uniform Fourier Transform to reach feasible reconstruction times. In addition, low-rank subspace projection is often used to reduce computational burden. While many implementations of NUFFT currently exists, they are not optimized for this kind of problems. Here, we propose a fast and memory-efficient NUFFT operator with embedded low-rank subspace projection. We demonstrate an order of magnitude of speed-up with comparable image quality compared to other high-level implementations.
|
||
2772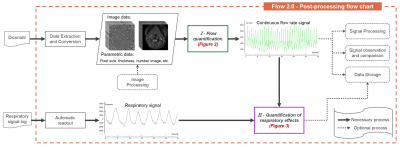 |
40 | Flow 2.0 - a flexible, scalable, cross-platform post-processing software for real-time phase contrast sequences
Liu Pan1,2, Sidy Fall1, and Olivier Baledent1,2
1CHIMERE UR 7516, Jules Verne University, Amiens, France, 2Medical Image Processing Department, Jules Verne University Hospital, Amiens, France Real-time phase contrast sequences (RT-PC) has potential value as a scientific and clinical tool in quantifying the effects of respiration on cerebral circulation. To simplify its complicated post-processing process, we developed Flow 2.0 software, which provides a complete post-processing workflow including converting DICOM data, image segmentation, image processing, data extraction, background field correction, anti-aliasing filter, signal processing and analysis and a novel time-domain method for quantifying the effect of respiration on the cerebral circulation. This end-to-end software allows us to quickly, robustly and accurately perform batch process RT-PC and multivariate analysis of the effects of respiration on cerebral circulation. |
||
2773 |
41 | A Cloud-based MR Spectroscopy Tool for Basis Set Simulation
Steve C.N. Hui1,2, Muhammad G. Saleh3, Helge J. Zöllner1,2, Georg Oeltzschner1,2, Hongli Fan1,4, Yue Li5, Yulu Song1,2, Hangyi Jiang1, Jamie Near6,7, Susumu Mori1,2, Hanzhang Lu1,2, and Richard A.E. Edden1,2
1Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 4Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5AnatomyWorks, LLC, Ellicott City, MD, United States, 6Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada, 7Sunnybrook Research Institute, Toronto, ON, Canada
MRSCloud is an online-based spectral simulation tool for brain metabolites. Up to 32 metabolites can be simulated to generate a basis set for linear combination modeling. The 1D projection method, coherence pathway filters and pre-calculation of propagators have been implemented for density-matrix simulations to run on a high-performance cloud server. Results indicated simulations were comparable with FID-A and difference between simulations of spatial points (21x21/41x41/101x101) were small. It allows community users to generate vendor, sequence, editing experimental-specific basis sets that are appropriate for their studies.
|
||
2774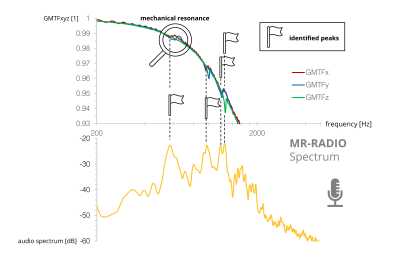 |
42 | Mechanical Resonance Recording and Analysis using auDIO files (MR-RADIO)
Hannes Dillinger1, Eva Peper1, Christian Guenthner1, and Sebastian Kozerke1
1ETH and University Zurich, Zurich, Switzerland
This work presents a web-based framework for the identification of mechanical resonances of MRI gradients systems based exclusively on mobile phone audio recordings. frequencies are used to guide beneficial combinations of TE/TR for PC-MRI and readout bandwidth settings for EPI sequences. Results demonstrate that the background phase in PC-MRI as well as beat phenomena during EPI readouts can be reduced. The web app can be found at https://mr-radio.herokuapp.com and its source code is publicly available at https://github.com/hdillinger/mr-radio in the spirit of reproducible research.
|
||
2775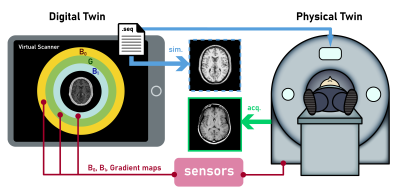 |
43 | Virtual Scanner 2.0: enabling the MR digital twin
Gehua Tong1, John Thomas Vaughan, Jr. 2, and Sairam Geethanath2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States Virtual Scanner is an open-source MR platform for end-to-end simulation and serves as a digital twin for real MR scanners in hardware and software aspects. In this update, we present new functions including RF pulse design and simulation, hardware-incorporated Bloch simulation that takes into account B0 and B1- fields, and non-Cartesian reconstruction. Three Google Colab notebooks are provided to demonstrate these functions and enable users to perform virtual experiments in a zero-footprint manner. Furthermore, we demonstrate the digital twin concept in a development iteration case using the SPGR sequence. |
||
2776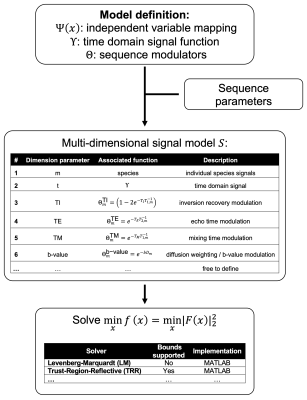 |
44 | ALFONSO: A versatiLe Formulation fOr N-dimensional Signal mOdel fitting of MR spectroscopy data and its application in MRS of body lipids
Stefan Ruschke1 and Dimitrios C. Karampinos1,2
1Department of Diagnostic and Interventional Radiology, Technical University of Munich, Munich, Germany, 2Munich Institute of Biomedical Engineering, Technical Universit of Munich, Munich, Germany
Multi-dimensional MR spectroscopy is frequently used for the probing of MR properties including the characterization of the water–fat environment in body applications. Previously, many studies used an ad-hoc multi-step approach where the fitting of the spectral content and the signal modelling was performed in separate steps albeit the theoretically compromised precision when compared to a joint fitting and modelling approach. ALFONSO allows the convenient and yet flexible definition of joint fitting and modelling strategies. Its utility and supremacy are demonstrated for the quantification of fat fraction in the liver, ADC in bone marrow and lipid droplet size in a phantom
|
||
2777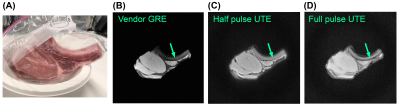 |
45 | An open-source repository of minimum echo time sequences
Gehua Tong1, John Thomas Vaughan, Jr. 2, and Sairam Geethanath2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States Ultrashort Echo Time (UTE) sequences are clinically useful in short T2 tissues such as bone, cartilage, and lung parenchyma. To help harmonize UTE protocols across clinical sites, we present an open-source repository implementing three basic radial minimal TE sequences using PyPulseq: 2D UTE, 3D UTE, and COncurrent Dephasing and Excitation (CODE). Functions for customized sequences and reconstruction scripts were provided for a complete pipeline. Echo Times (TEs) from 0.12 to 0.82 ms were achieved, and 27.6% more signal was captured in the bone and connective tissue in 2D ex vivo images compared to the GRE reference. |
||
2778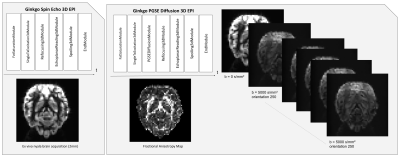 |
46 | Ginkgo: a novel modular and and Open Source MRI pulse sequence development framework dedicated to MRI systems
Anaïs Artiges1, Franck Mauconduit1, Ivy Uszynski1, Baptiste Mulot2, Elodie Chaillou3, Philippe Ciuciu4, and Cyril Poupon1
1BAOBAB, NeuroSpin, Paris-Saclay University, CNRS, CEA, Gif-sur-Yvette, France, 2Beauval Nature, ZooParc de Beauval, Saint-Aignan, France, 3UMR 85 Physiologie de la Reproduction et des Comportements, INRAE Centre Val-de-Loire, Nouzilly, France, 4Parietal, NeuroSpin, Paris-Saclay University, Inria, CEA, Gif-sur-Yvette, France
We created a new object-oriented environment for MR pulse-sequence development based on IDEA VE11C and above versions using an Open Science philosophy. This Ginkgo toolkit uses a modular structure to facilitate the design of pulse sequences using the aggregation of basic open-source sequence blocks available from the toolkit. Proofs of concept of the productivity gain reached using Ginkgo are provided through the implementation of a series of sequence models including a diffusion-weighted PGSE 3D EPI sequence.
|
||
2779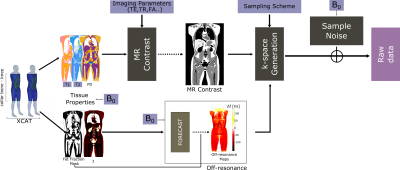 |
47 | Realistic Simulation of Body Composition MRI and Field-Strength Dependence
Ecrin Yagiz1, Sophia X. Cui2, Nam G. Lee1, Bilal Tasdelen1, Ye Tian1, and Krishna S. Nayak1,3
1Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Siemens Medical Solutions USA, Inc.,, Los Angeles, CA, United States, 3Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States
MRI is used extensively for body composition assessment in large part due to the increasing prevalence of metabolic disease and obesity. In this work, we provide a framework for simulating body composition MRI, including non-Cartesian sequences and emerging low field strengths. This framework can be utilized to investigate the optimality of acquisition parameters (e.g., resolution, TE, sampling trajectory) and the performance of fat/water separated reconstruction algorithms.
|
||
2780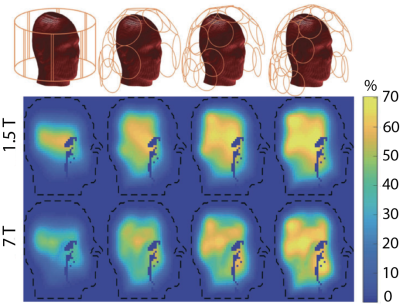 |
48 | A software tool to assess radiofrequency coil designs with respect to ultimate intrinsic performance limits
Eros Montin1, Ioannis P Georgakis1,2, Bei Zhang3, and Riccardo Lattanzi1,4
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 2Corsmed, Stockolm, Sweden, 3Advanced Imaging Research Center, UT Southwestern Medical Center, Dallas, TX, United States, 4Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine,, New York, NY, United States
This work introduces POIROT (Performance Observer In Receive or Transmit), a web-based tool for the assessment of receive and transmit coil designs against ultimate intrinsic signal-to-noise ratio and transmit efficiency, respectively. It enables engineers, for the first time, to evaluate how good is a design and whether there is further room for improvement before building a prototype. POIROT currently includes ultimate intrinsic data for three numerical head models at different resolutions and magnetic field strengths. POIROT could be integrated with rapid numerical EM modeling tools to develop a pipeline for coil design optimization that uses ultimate performance as the benchmark.
|
||
2781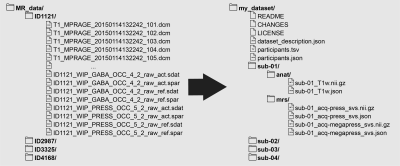 |
49 | A BIDS extension proposal for magnetic resonance spectroscopy data
Mark Mikkelsen1, Dickson Wong2, Wolfgang Bogner3, Yaroslav O. Halchenko4, Damon G. Lamb5,6,7,8,9, Paul G. Mullins10, Georg Oeltzschner11,12, and Martin Wilson13
1Department of Radiology, Weill Cornell Medicine, New York, NY, United States, 2Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, 3High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Psychological and Brain Sciences, Dartmouth College, Hanover, NH, United States, 5Departments of Psychiatry, Neuroscience, and Biomedical Engineering, University of Florida, Gainesville, FL, United States, 6Center for OCD, Anxiety, and Related Disorders, University of Florida, Gainesville, FL, United States, 7Center for Cognitive Aging and Memory, University of Florida, Gainesville, FL, United States, 8McKnight Brain Institute, University of Florida, Gainesville, FL, United States, 9Brain Rehabilitation Research Center, Malcom Randall Veterans Affairs Medical Center, Gainesville, FL, United States, 10Bangor Imaging Unit, School of Psychology, Bangor University, Bangor, United Kingdom, 11Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 12F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 13Centre for Human Brain Health and School of Psychology, University of Birmingham, Birmingham, United Kingdom
Magnetic resonance spectroscopy (MRS) studies can generate extensive and complex datasets. Datasets can be organized idiosyncratically across (and even within) labs. However, sharing data and reproducing results without the added challenge of parsing lab-specific file organization is paramount to open science. Sharing of MRS data that is independent of lab-specific practices would greatly aid transparency and reusability. Here, we present an extension to the Brain Imaging Data Structure (BIDS) specification for MRS data. MRS-BIDS adopts the established tenets of standardization of BIDS and applies them to MRS data while considering the nuances relevant to MRS methodology.
|
||
2782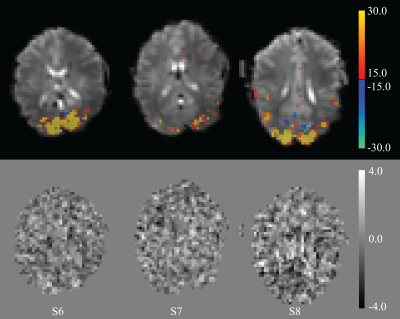 |
50 | Study of potential immediate effects of neuronal activity on the MRI signal in humans
Peter van Gelderen1, Jacco A de Zwart1, and Jeff H Duyn1
1Advanced MRI section, LFMI, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States A recent study has shown rapid MRI signals in response to whisker pad stimulation in mice that track electrical activity. We used rapid, high SNR imaging at 7 T to look for a possible direct effect of visual stimulation on MRI signal in human visual cortex. Two rapid stimulation protocols and variation of T1-weighting failed to generate observable signal changes above the 0.01% detection threshold, well below the previously reported effect size of 0.15%. This confirms the well-established difficulty in developing more direct measures of neuronal activity than available with BOLD-fMRI. |
||
2783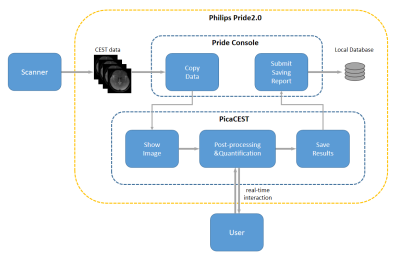 |
51 | A Software Tool for Inline Post-processing and Visualization of Chemical Exchange Saturation Transfer (PV-CEST) on 3T Scanner
Chuyu Liu1, Yishi Wang2, Zhensen Chen3, Yajing Zhang4, and Xiaolei Song1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Philips Healthcare, Beijing, China, 3Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 4MR Clinical Science, Philips Healthcare, Suzhou, China
We developed a real-time software for CEST post-processing and visualization on Philips 3T scanner, termed as PV-CEST. PV-CEST runs in the vendor provided PRIDE 2.0 environment, providing image view, Z-spectra analysis and quantitative maps calculation functionalities during scanning procedure, which could have great clinical utility.
|
||
2784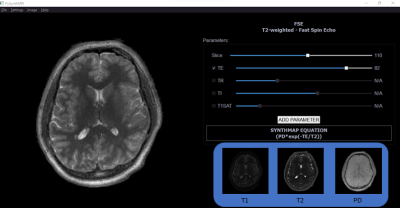 |
52 | PySynthMRI: An open-source Python tool for Synthetic MRI
Luca Peretti1,2,3, Matteo Cencini3, Paolo Cecchi2,4, Graziella Donatelli2,4, Mauro Costagli3,5, and Michela Tosetti3
1University of Pisa, Pisa, Italy, 2Imago7 Research Foundation, Pisa, Italy, 3IRCCS Stella Maris, Pisa, Italy, 4Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy, 5University of Genoa, Genoa, Italy
Synthetic MR imaging allows reconstruction of different image contrasts from a single acquisition, reducing scan times. Here we introduce PySynthMRI, an open-source tool which provides a user-friendly and fluid interface to synthesize and modify contrast-weighted images. The tool allows the user to add and adjust virtual scanner parameters showing, in real time, how the resulting synthesized images vary accordingly, which makes PySynthMRI a valuable tool for both research and teaching. The generated images can be exported in the preferred format.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.