Digital Poster
DSC & DCE Perfusion
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1579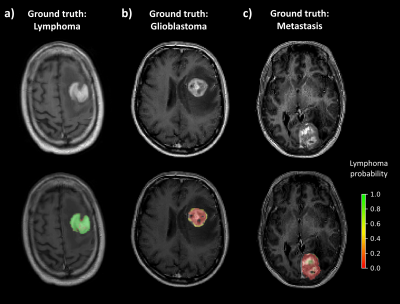 |
34 | Deep learning voxelwise classification of primary central nervous system lymphoma using DSC-PWI normalized time-intensity curves
Alonso Garcia-Ruiz1, Albert Pons-Escoda2, Francesco Grussu1, Pablo Naval-Baudin2, Camilo Monreal1, Antonio Lopez-Rueda3, Laura Oleaga3, Carlos Majos2, and Raquel Perez-Lopez1
1Vall d'Hebron Institute of Oncology, Barcelona, Spain, 2Bellvitge University Hospital, L'Hospitalet de Llobregat, Spain, 3Hospital Clínic de Barcelona, Barcelona, Spain
The robustness and accuracy of current MR methods to differentiate brain tumours is limited. In this study we investigate the potential of dynamic susceptibility perfusion-weighted imaging (DSC-PWI) normalized time-intensity-curves (nTIC) to support lymphoma diagnosis by harnessing voxelwise and temporal information to train a convolutional neural network (CNN). This novel approach discriminated patients with lymphoma from glioblastoma and metastasis with an average accuracy of 0.94, using only a limited number of patients for training, outperforming standard DSC-PWI measurements. Furthermore, it provides voxel-by-voxel lymphoma probability maps to further help visual diagnosis of neuroradiologists.
|
||
1580 |
35 | DCE-MRI kinetic models for measuring subtle blood-brain barrier leakage– the importance of modelling finite interstitial volume fraction
Martin Kozár1,2, Sarah Al-Bachari3, Geoff J. M. Parker4,5, Laura M. Parkes1,2, and Ben R. Dickie2,6
1Division of Neuroscience and Experimental Psychology, The University of Manchester, Manchester, United Kingdom, 2Geoffrey Jefferson Brain Research Centre, The University of Manchester, Manchester, United Kingdom, 3Faculty of Health and Medicine, The University of Lancaster, Lancaster, United Kingdom, 4Centre for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 5Bioxydyn Limited, Manchester, United Kingdom, 6Division of Informatics, Imaging and Data Science, The University of Manchester, Manchester, United Kingdom
The Patlak model is commonly applied to brain DCE-MRI to quantify subtle blood-brain barrier permeability. This is based on existing evidence showing the Patlak model provides the optimal fit compared to other candidate models, including the extended Tofts model (Heye, Thrippleton et al. 2016). In this study, we compared five different tracer kinetic models in data with higher temporal resolution and determined that models which assume finite interstitial volume (ve) (e.g. extended Tofts) consistently outperform models that assume infinite ve (e.g. Patlak), particularly in gray matter.
|
||
1581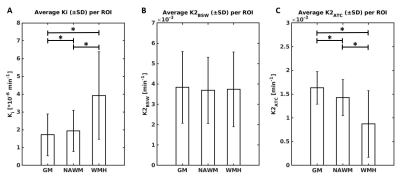 |
36 | To what extent is DSC-MRI able to detect subtle blood-brain barrier leakage in cerebral small-vessel disease?
Elles Elschot1,2, Walter Backes1,2,3, Joost de Jong1,2, Gerhard Drenthen1,2, Sau May Wong1, Julie Staals3,4, Robert van Oostenbrugge2,3,4, Rob Rouhl4, and Jacobus Jansen1,2,5
1Radiology and Nuclear Medicine, Maastricht University Medical Center +, Maastricht, Netherlands, 2School for Mental Health and Neuroscience, Maastricht University, Maastricht, Netherlands, 3CARIM School for Cardiovascular Diseases, Maastricht University, Maastricht, Netherlands, 4Neurology, Maastricht University Medical Center +, Maastricht, Netherlands, 5Electrical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands
This study investigated to what extent blood-brain barrier (BBB) leakage (1) can be measured with DSC-MRI and (2) is influenced by perfusion. In vivo DCE (golden-standard) and DSC data of patients with cerebral small vessel disease (cSVD) and elderly controls were used, as well as simulations of signal curves. DSC-MRI, in contrast to DCE-MRI, is not sensitive enough to measure subtle leakage in cSVD. Further research is required to better disentangle perfusion effects from leakage, and therefore correction methods should be used with caution for measuring subtle leakage.
|
||
1582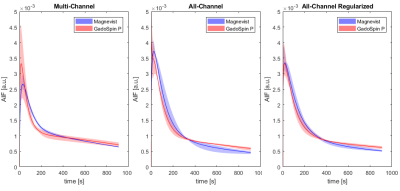 |
37 | Arterial Input Function Estimation Using All-Channel Blind Deconvolution with Spatial Regularization in DCE-MRI
Radovan Jirik1, Michal Bartos2, Ondrej Macicek1, and Zenon Starcuk, jr.1
1Institute of Scientific Instruments, Czech Academy of Sciences, Brno, Czech Republic, 2Institute of Information Theory and Automation, Czech Academy of Sciences, Praha, Czech Republic
This work is focused on estimation of an arterial input function (AIF) in DCE-MRI. We propose a new concept of blind deconvolution to use as much available information as possible. In contrast to multi-channel blind deconvolution based on processing of selected tissue signals, our all-channel blind deconvolution uses tissue signals of all voxels within the studied tissue to simultaneously estimate the AIF and perfusion maps. In addition, edge-preserving spatial regularization is included in the perfusion-map estimation scheme. The method is illustrated on DCE-MRI recordings of tumor-bearing mice.
|
||
1583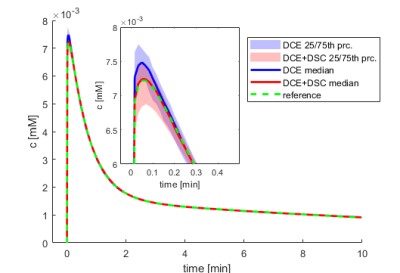 |
38 | Combining DCE and DSC-MRI in Arterial Input Function Estimation Using Multi-Channel Blind Deconvolution
Ondrej Macicek1, Radovan Jirik1, Peter Latta2, and Zenon Starcuk jr.1
1Institute of Scientific Instruments of the Czech Academy of Sciences, Brno, Czech Republic, 2Central European Institute of Technology, Masaryk University, Brno, Czech Republic
We propose to combine DCE and DSC-MRI in a simultaneous acquisition and processing scheme to provide more information for estimation of the arterial input function using multi-channel blind deconvolution. On simulated and flow-phantom data, we show that DCE-DSC blind deconvolution is superior to standard DCE blind deconvolution, especially for low signal-to-noise-ratio conditions.
|
||
1584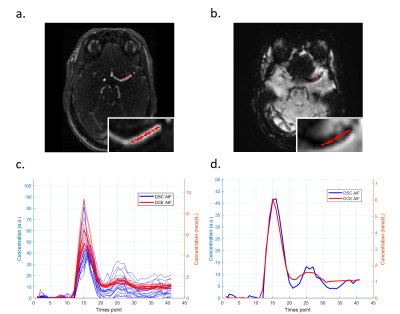 |
39 | Comparison of arterial input functions obtained through back-to-back acquisition of DCE and DSC MRI
Chih-Hsien Tseng1, Jaap Jaspers2, Alejandra Mendez Romero2, Piotr Wielopolski3, Matthias van Osch 4, and Frans Vos1,3
1Imaging Physics, Delft University of Technology, Delft, Netherlands, 2Radiation Oncology, Erasmus Medical Center, Rotterdam, Netherlands, 3Radiology, Erasmus Medical Center, Rotterdam, Netherlands, 4Radiology, Leiden University Medical Center, Leiden, Netherlands We aimed to compare the AIF determined from DCE-MRI with the AIF from DSC-MRI in back-to-back imaging, using the same dose of Gadobutrol. The DCE-driven AIFs showed the same ‘pattern’ as the DSC-driven AIFs, but had smaller variance of the shape and peak value. A linear relation was found between the contrast agent concentration from DCE and ΔR2*, but the linear coefficients showed large variation across regions and subjects. Our findings show that the DCE-driven AIF has the potential to serve as an alternative for the DSC-driven AIFs for quantitative perfusion assessment. |
||
1585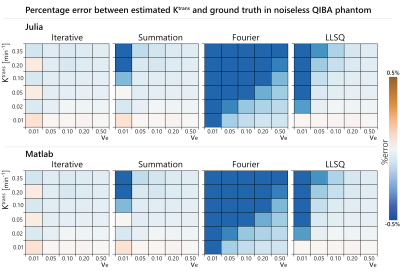 |
40 | Accuracy of quantitative DCE-MRI depends on the implementation of the convolution operation
Zaki Ahmed1,2 and Ives R. Levesque2,3
1Department of Radiology, Mayo Clinic, Rochester, MN, United States, 2Medical Physics Unit, McGill University, Montreal, QC, Canada, 3Research Institute of the McGill University Health Centre, Montreal, QC, Canada
Quantitative DCE-MRI analysis requires fitting a model to the acquired enhancing time-course data. These models involve a convolution operation, which has a variety of possible implementations. The goal of this work was to evaluate three different implementations of the convolution operation: (i) Fourier, (ii) Summation, (iii) Iterative. The accuracy and execution time of each implementation was compared on a virtual phantom. It was found that the iterative technique was the fastest while also having the best overall accuracy.
|
||
1586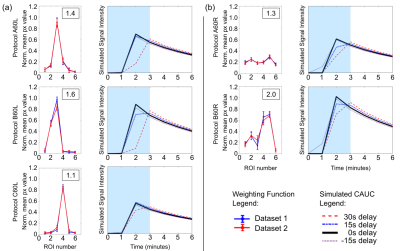 |
41 | The effect of k-space sampling patterns on contrast agent uptake curves in dynamic contrast-enhanced MRI of the breast
Owen Alun White1, Evanthia Kousi1, Clarrissa Sanders2, and Maria Angelica Schmidt1
1Radioisotope & Imaging Physics, The Royal Marsden NHS Foundation Trust, London, United Kingdom, 2St George's University Hospital NHS Foundation Trust, London, United Kingdom
Here we propose a novel method to assess the effect of different k-space sampling patterns on contrast agent uptake curves (CAUCs), which does not require specialist equipment. These effects on CAUCs are often overlooked, and must be addressed in the context of the many different national and international recommendations, and the prominence given to abbreviated DCE protocols. We validated the method to assess different k-space sampling patterns and used it to simulate the effect on contrast-agent uptake curves. We show that conventional protocols acquiring each frame in approximately 60s have more robust CAUC classification than abbreviated protocols.
|
||
1587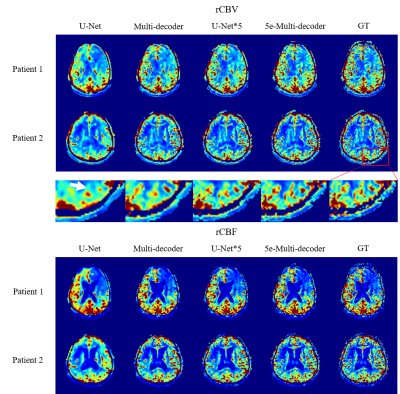 |
42 | Acquiring perfusion maps from contrast-enhanced MRA using deep learning approaches
Muhammad Asaduddin1, Hong Gee Roh2, Hyun Jeong Kim3, Eung Yeop Kim4, and Sung-Hong Park1
1Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Konkuk University Medical Center, Seoul, Korea, Republic of, 3Department of Radiology, Daejeon St. Mary's Hospital, The catholic University of Korea, Daejeon, Korea, Republic of, 4Department of Radiology, Gil Medical Center, Gachon University College of Medicine, Incheon, Korea, Republic of
Perfusion maps and dynamic angiograms are both important for stroke/tumor treatment but commonly acquired in separate scans and thus may require additional injection of contrast agent for best results. In this work, we present a deep learning method to acquire perfusion maps from contrast-enhanced MRA data. Our results showed that an architecture of multiple decoders and an enhanced encoder produced perfusion maps that were visually and quantitatively similar to the standard DSC MRI-based perfusion maps. This approach enables us to acquire accurate perfusion maps and angiogram using a single contrast agent injection, reducing costs and risks while improving patient comfort.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.