Digital Poster
Deep Learning Image Reconstruction I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
0841 |
66 | Accelerated MRI Thermometry with Multi-Echo Multi-Slice GRE
Yuval Zur1, Itzhak Pinhas1, and Boaz Shapira1
1Insightec Ltd, Haifa, Israel
An accelerated multi echo GRE sequence is presented for Focused Ultrasound Thermometry. An under sampled k space dataset is acquired during heating, to reduce scan time and enable multi-slice acquisition. Prior to heating, a fully sampled baseline image is acquired. Based on the baseline image, an iterative reconstruction algorithm fills-in the missing lines of each hot image. Accelerations of up to x15 with 16 echoes enable acquisition of 5 – 10 slices in 3.5 sec. Real-time reconstruction of 800 msec/5 slices was implemented on a FUS-MRI machine.
|
||
0842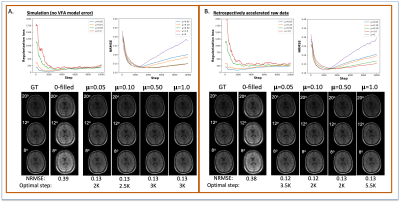 |
67 | An untrained deep learning method with model-based regularization for reconstructing dynamic MR images from retrospectively accelerated data
Kalina P Slavkova1, Julie C DiCarlo2,3, Viraj Wadhwa4, Thomas E Yankeelov2,3,5,6,7, and Jonathan I Tamir2,4,6
1Department of Physics, The University of Texas at Austin, Austin, TX, United States, 2Oden Institute for Computational Engineering and Sciences, The University of Texas at Austin, Austin, TX, United States, 3Livestrong Cancer Institutes, The University of Texas at Austin, Austin, TX, United States, 4Department of Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States, 5Department of Biomedical Engineering, The University of Texas at Austin, Austin, TX, United States, 6Department of Diagnostic Medicine, The University of Texas at Austin, Austin, TX, United States, 7Department of Oncology, The University of Texas at Austin, Austin, TX, United States
Acquiring high-resolution MRI data for tissue parameter mapping for quantitative imaging requires additional scan time. As a proof-of-principle, we evaluated the ability of the ConvDecoder architecture regularized with a physical model to reconstruct accelerated variable-flip angle MRI data of the brain for T1-mapping. The performance of our method was compared to non-regularized ConvDecoder, low rank reconstruction, and compressed sensing. Our results suggest that ConvDecoder with physics-based regularization may provide a stopping condition for training that is not dependent on the ground truth data while improving parameter mapping at higher accelerations.
|
||
0843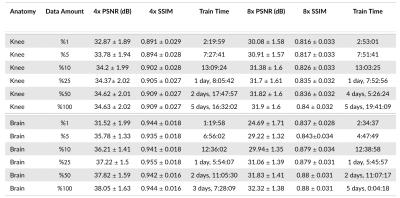 |
68 | Systematic Standardization of Deep Learning Based Accelerated MRI Reconstruction Pipelines
Beliz Gunel1,2, Arjun Divyang Desai1,2, Shreyas Vasanawala3, Akshay Chaudhari3,4, and John Pauly1
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Equal contribution, Stanford University, Stanford, CA, United States, 3Radiology, Stanford University, Stanford, CA, United States, 4Biomedical Data Science, Stanford University, Stanford, CA, United States
Deep learning based accelerated MRI reconstruction pipelines have potential to enable higher acceleration factors compared to traditional methods with fast reconstruction times and improved image quality. Although there have been studies regarding model architecture, loss function, and k-space undersampling patterns; the effect of scanner parameters, variations in sensitivity map estimation, training data requirement, and engineering decisions during model optimization and evaluation on the reconstruction performance remain largely unexplored. We systematically study the impact of such and show that such data extent, re-processing, and metric computation impact performance to the same or at larger extents than new architectures and loss functions.
|
||
0844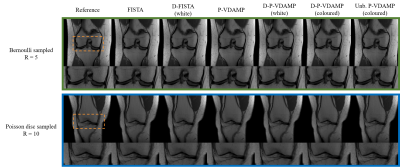 |
69 | Deep Plug-and-Play multi-coil compressed sensing MRI with matched aliasing: the Denoising-P-VDAMP algorithm
Charles Millard1, Aaron Hess1, Boris Mailhe2, and Jared Tanner3
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Siemens, Princeton, NJ, United States, 3Mathematical Institute, University of Oxford, Oxford, United Kingdom
We present the Denoising Parallel Variable Density Approximate Message Passing (D-P-VDAMP) algorithm for multi-coil compressed sensing MRI with a learned prior. To our knowledge, D-P-VDAMP is the first Plug-and-Play method for multi-coil k-space data where the distribution of the training data's aliasing matches the actual distribution seen during reconstruction. We evaluate the performance of the proposed method on the fastMRI knee dataset and find substantial improvements in reconstruction quality compared with Plug-and-Play FISTA with the same network architecture in similar training and reconstruction time.
|
||
0845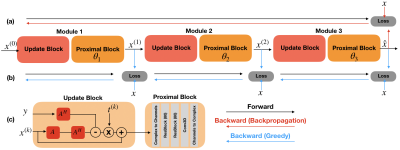 |
70 | Parallel Greedy Learning for Accelerating Cardiac Cine MRI
Batu M Ozturkler1, Arda Sahiner1, Tolga Ergen1, Arjun D Desai1, Christopher M Sandino1, Shreyas Vasanawala2, John M Pauly1, Morteza Mardani1, and Mert Pilanci1
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Deep unrolled models have recently shown state-of-the-art performance for reconstruction of dynamic MR images. However, training these networks via backpropagation is limited by intensive memory and compute requirements to calculate gradients and store intermediate activations per layer. Motivated by these challenges, we propose an alternative training method by greedily relaxing the training objective. Our approach splits the end-to-end network into decoupled network modules, and optimizes each module separately, avoiding the need to compute end-to-end gradients. We demonstrate that our method outperforms end-to-end backpropagation by 3.3 dB in PSNR and 0.025 in SSIM with the same memory footprint.
|
||
0846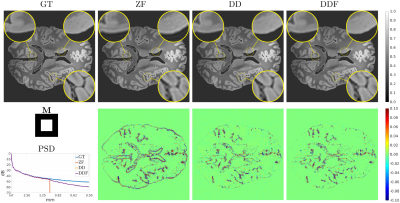 |
71 | Deep generative MRI reconstruction for unsupervised Gibbs ringing correction
Lucilio Cordero-Grande1,2, Enrique Ortuño-Fisac1, Jonathan O'Muircheartaigh2,3, Jo Hajnal2, and María Jesús Ledesma-Carbayo1
1Biomedical Image Technologies, ETSI Telecomunicación, Universidad Politécnica de Madrid & CIBER-BBN, Madrid, Spain, 2Centre for the Developing Brain & Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 3Department for Forensic and Neurodevelopmental Sciences, Institute of Psychiatry, Psychology and Neuroscience & MRC Centre for Neurodevelopmental Disorders, King's College London, London, United Kingdom
MRI reconstruction is formulated as the retrieval of the parameters of a deep decoder network fitted to the observations by an image formation model including the truncation of high-frequency information. Solutions without Gibbs ringing at any prescribed image grid can be obtained naturally by the model without training or ad-hoc post-corrections. We present quantitative and visual results for spectral extrapolation of magnitude images at different scales in an in-silico experiment and a high resolution ex-vivo brain MRI scan. After minor modifications to deal with complex data, the architecture is applied to 2D parallel imaging showing promising visual results.
|
||
0847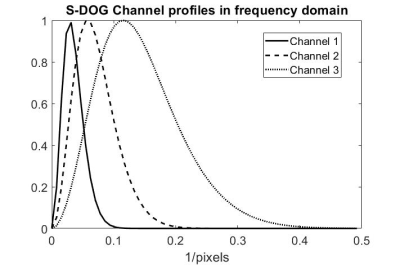 |
72 | Evaluation of Neural Network Reconstruction of Undersampled Data using a Human Observer Model of Signal Detection
Joshua D Herman1, Marcus L Wong1, Sajan G Lingala2, and Angel R Pineda1
1Mathematics Department, Manhattan College, Riverdale, NY, United States, 2Roy J. Carver Department of Biomedical Engineering, University of Iowa, Iowa City, IA, United States
We evaluated images from undersampled data using a U-Net with common metrics (SSIM and NRMSE) and with a model for human observer detection, the sparse difference-of-Gaussians (S-DOG). We also studied how the results vary when changing the loss function and training set size. We saw that the S-DOG model would choose an undersampling of 2X while SSIM and NRMSE would choose 3X. In previous work, human observers also chose a 2X acceleration. The S-DOG model led to the same conclusion as the human observers. This result was consistent with changes in training set size and loss function.
|
||
0848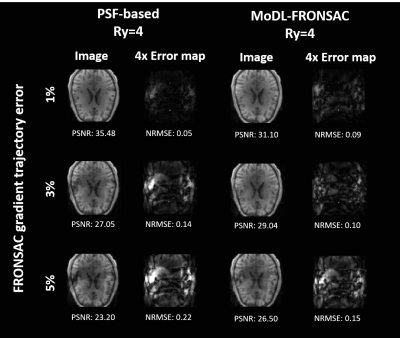 |
73 | Deep Learning Reconstruction for FRONSAC
Zhehong Zhang1 and Gigi Galiana2
1Department of Biomedical Engineering, Yale University, New Haven, CT, United States, 2Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
This work is the first to apply deep learning to the reconstruction of images encoded with nonlinear gradients. We apply a model-based deep learning network (MoDL) to simulated FRONSAC images and compare these to a PSF-based matrix inversion as well as cg-SENSE. The MoDL based reconstruction did not significantly change the behavior of signal noise. However, results demonstrate that the model-based deep learning network can outperform traditional reconstruction methods at high undersampling factors. Simulations also suggests that the regularizing network has potential to correct for miscalibration in the nonlinear gradient trajectory.
|
||
0849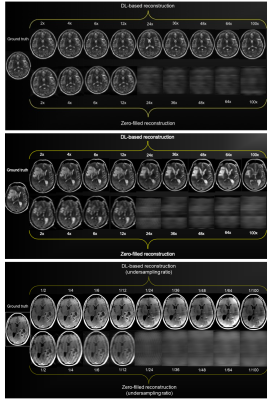 |
74 | Properties of 2D MR image reconstructions with deep neural networks at high acceleration rates
Matthew J. Muckley1, Tullie Murrell2, Alireza Radmanesh3, Florian Knoll4, Zhengnan Huang3, Anuroop Sriram5, Daniel K. Sodickson3, and Yvonne W. Lui3
1Facebook AI Research, New York, NY, United States, 2shaped.ai, New York, NY, United States, 3Radiology, NYU School of Medicine, New York, NY, United States, 4Artificial Intelligence in Biomedical Engineering, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany, 5Facebook AI Research, Menlo Park, CA, United States
We assess the properties of deep neural network-reconstructed brain MR images in the high acceleration regime at factors up to 100. We have three contributions: 1) metrics on model performance from 2- to 100-fold accelerations, 2) a Monte Carlo procedure for scoring the quality of model reconstructions using only subsampled data, and 3) assessment of the acceleration effects on pathology in six cases. Our Monte Carlo procedure can estimate ground truth PSNR with coefficients of determination greater than 0.5 using only subsampled data. Our pathology results were stable in DNN reconstructions up to 8-fold acceleration.
|
||
0850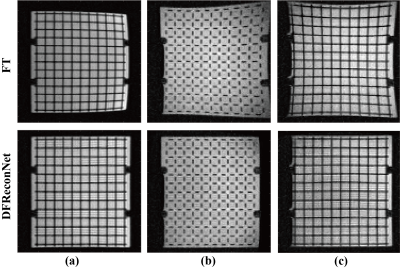 |
75 | Distortion Free Image Reconstruction using a Deep Neural Network for an MRI-Linac
Shanshan Shan1,2, Yang Gao3, Paul Liu1,2, Brendan Whelan1, Hongfu Sun3, Feng Liu3, Paul Keall1,2, and David Waddington1,2
1ACRF Image X Institute, University of Sydney, Sydney, Australia, 2Ingham Institute For Applied Medical Research, Sydney, Australia, 3School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
The advent of MRI-guided radiotherapy has elevated demand for high geometric fidelity imaging. However, gradient nonlinearity can cause image distortion, which limits the accuracy of radiotherapy. In this work, we develop a deep neural network, namely DFReconNet, to reconstruct distortion free images directly from raw k-space in real time. Experiments on simulated brain datasets and phantom images acquired from an MRI-Linac demonstrated the utility of the proposed method.
|
||
0851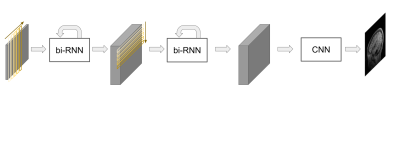 |
76 | A deep learning based direct mapping method for EPI image reconstruction
Changheun Oh1, Sung Suk Oh2, Jun-Young Chung1, and Yeji Han1
1Gachon University, Incheon, Korea, Republic of, 2Daegu Gyeongbuk Medical Innovation Foundation, Daegu, Korea, Republic of
Echo planar imaging (EPI) is one of the fastest MRI pulse sequences that can acquire whole k-space data during a single excitation. In EPI, partial acquisition can be used for reducing the TR and the effective TE can be further reduced as the portion of the acquired k-space is reduced. However, the quality of the image decreases drastically as the acquired k-space portion is reduced. In this study, we proposed a solution for image reconstruction from partially acquired EPI based on a deep-learning based direct mapping method, where the images are directly reconstructed from input k-space data.
|
||
0852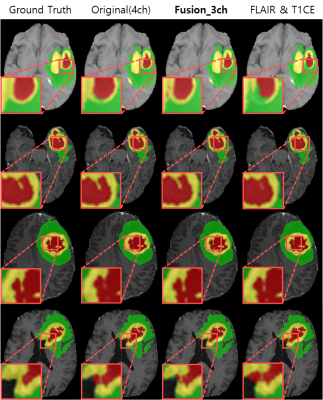 |
77 | Brain Tumor Segmentation Using 3D CMM-Net with Limited and Accessible MR Images
Yoonseok Choi1, Mohammed A. Al-masni1, and Dong-Hyun Kim1
1Department of Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of
In this work, we investigate the possibility of using a limited number of medical MR images for brain tumor segmentation. This can assist to perform the segmentation task using limited and accessible data in clinical practice. We observed that the FLAIR and T1CE are the most suitable images that maintain comparable segmentation performances similar to the case of using more data (i.e., T1, T2, FLAIR, and T1CE). To further improve the segmentation results, we augment the selected image pair and generate an additional fusion map using the Singular Value Decomposition (SVD).
|
||
0853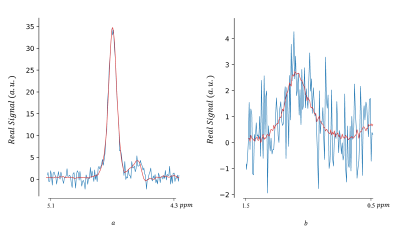 |
78 | Magnetic Resonance Spectroscopic Imaging Data Denoising by Manifold Learning: An Unsupervised Deep Learning Approach
Amirmohammad Shamaei1,2, Jana Starčuková1, Radim Kořínek1, and Zenon Starčuk Jr1
1Institute of Scientific Instruments of the Czech Academy of Sciences, Brno, Czech Republic, 2Department of Biomedical Engineering, Faculty of Electrical Engineering and Communication, Brno University of Technology, Brno, Czech Republic
This work demonstrates an unsupervised deep-learning approach to MRS(I) data denoising, incorporating a non-linear model without relying on MR-theoretical physical prior knowledge. The implemented autoencoder learns the underlying low-dimensional manifold in high-dimensional data vectors representing MRS(I) voxels. The method, developed for denoising water-fat spectroscopic images, was tested against both numerical and acquired phantoms containing milk cream. The proposed method shows results comparable with other techniques in boosting SNR and has been found more robust in low concentration component denoising. Deep learning data denoising for MRSI might result in faster acquisition, vital for MRSI clinical application.
|
||
0854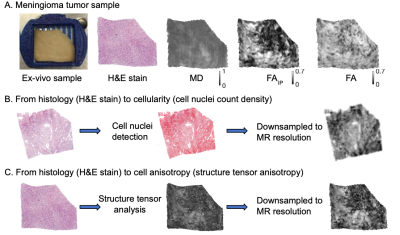 |
79 | Explaining variation in DTI parameters with meningioma microscopy: A comparison between a neural network and an image-feature-based approach
Jan Brabec1, Magda Friedjungová2, Daniel Vašata2, Elisabet Englund3, Linda Knutsson1,4,5, Filip Szczepankiewicz6, Pia C Sundgren6, and Markus Nilsson6
1Medical Radiation Physics, Lund University, Lund, Sweden, 2Department of Applied Mathematics, Faculty of Information Technology, Czech Technical University, Prague, Czech Republic, 3Division of Oncology and Pathology, Department of Clinical Sciences, Lund University, Skåne University Hospital, Lund, Sweden, 4Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 6Diagnostic Radiology, Clinical Sciences Lund, Lund University, Lund, Sweden
Diffusion MRI reflects tissue microstructure and mean diffusivity (MD) and fractional anisotropy (FA) are often readily interpreted in terms of cellularity and cell anisotropy, respectively. Here, we investigated to which degree histological features accounts for their variations in fixed sections of meningioma tumors. Histological slices were quantified in terms of cellularity and cell anisotropy, or by a neural network. Results show that in some cases the majority of the variation can be attributed to cellularity whereas in others none. Similarly, in some samples only a minority of the variability is attributable to the variability in FA explained by neural network.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.