Digital Poster
Quantitative Imaging I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
2521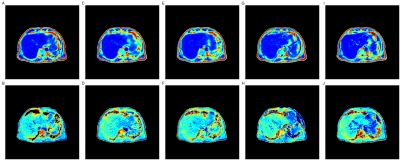 |
53 | Predicting PDFF and R2* from Magnitude-Only Two-Point Dixon MRI Using Generative Adversarial Networks Video Permission Withheld
Nicolas Basty1, Marjola Thanaj1, Madeleine Cule2, Elena P. Sorokin2, E. Louise Thomas1, Jimmy D. Bell1, and Brandon Whitcher1
1Research Centre for Optimal Health, University of Westminster, London, United Kingdom, 2Calico Life Sciences LLC, South San Francisco, CA, United States
We implement generative adversarial network (GAN) models to predict fully quantitative parameters from a complex-valued multiecho MRI sequence using only data from the magnitude-only two-point Dixon acquisition in the UK Biobank abdominal protocol. The training data consists of in- and opposed-phase channels from the Dixon sequence as inputs and the proton density fat fraction (PDFF) and R2* parameter maps estimated from the IDEAL acquisition as outputs. We compare conditional and cycle GANs, where the conditional GAN models outperformed the cycleGAN models in SSIM, PSNR and MSE. PDFF predictions were better than R2* predictions for all models.
|
||
2522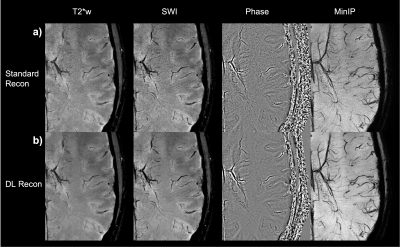 |
54 | Fast and High-Resolution T2* weighted and susceptibility-based MRI using 3D-EPI with Deep Learning Reconstruction
Brice Fernandez1, R. Marc Lebel2, Xinzeng Wang3, Arnaud Guidon4, Suchandrima Banerjee5, Stefan Skare6,7, and Tim Sprenger8
1GE Healthcare, Buc, France, 2GE Healthcare, Calgary, AB, Canada, 3GE Healthcare, Houston, TX, United States, 4GE Healthcare, Boston, MA, United States, 5GE Healthcare, Menlo Park, CA, United States, 6Karolinska University Hospital, Stockholm, Sweden, 7Karolinska Institutet, Stockholm, Sweden, 8GE Healthcare, Stockholm, Sweden Fast and high-resolution T2*w and susceptibility-weighted imaging is an essential part of brain MR assessment for many neurological conditions. Very high spatial resolution can be helpful for visualization of microvascular details as well as the central vein sign in white matter Multiple Sclerosis lesions, but such scans have a long acquisition time. In this abstract, we demonstrate high-quality whole-brain T2*w and susceptibility-based MRI at 0.5 mm isotropic resolution in less than 4 minutes using a 3D-echo-planar acquisition and a deep learning reconstruction. Finally, straightforward improvements are discussed for further reduction of the scan time. |
||
2523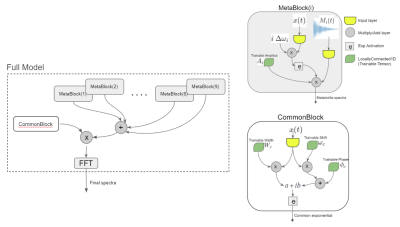 |
55 | Prior-knowledge MRS Metabolite Quantification using Deep Learning Frameworks: A proof-of-concept Video Permission Withheld
Federico Turco1 and Johannes Slotboom1
1SCAN, Institute of Diagnostic and Interventional Neuroradiology, Bern, Switzerland
We implemented a mathematical representation of a prior-knowledge model in a Neural Network using a Tensorflow. The trainables tensors are directly the free parameters of the model and we do metabolite quantification by overfitting the output to the signal that we want to replicate. We found that this way of fitting has a relatively low performance in time domain but similar to the state-of-the-art (TDFDFit) when using frequency domain. In addition we have a faster method and it can be used in future works as a component of a more complex network.
|
||
2524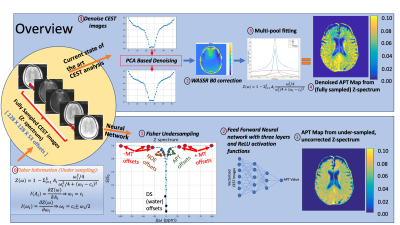 |
56 | Amide Proton Transfer (APT) Mapping from Undersampled Z-spectra in the Brain Using Deep Learning
Karandeep Cheema1,2, Pei Han1,2, Hui Han1, Yibin Xie1,2, Anthony Christodoulou1,2, and Debiao Li1,2
1Biomedical Imaging Research Institute, Cedars Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, University of California, Los Angeles (UCLA), Los Angeles, CA, United States
CEST Imaging requires radio frequency pulses at several frequency offsets to generate CEST maps (APT, NOE, DS, MT). In this study, we aimed to generate APT maps from CEST images of undersampled frequency offsets using deep learning, which can potentially reduce the total scan time of CEST imaging. The Z-spectrum was undersampled by a factor of 3.5 using the Fisher information gain analysis. Fitting results from the proposed method were compared with those from multi-pool fitting with fully sampled Z-spectrum. We have shown that it is feasible to reconstruct APT maps from undersampled, uncorrected CEST images.
|
||
2525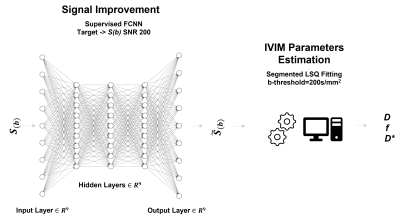 |
57 | Fully Connected Deep Neural Network combined with Segmented Least Square Fitting for improved extraction of IVIM Parameters
Alfonso Mastropietro1, Elisa Scalco1, Daniele Procissi2, Nicola Bertolino2, and Giovanna Rizzo1
1Istituto di Tecnologie Biomediche, Consiglio Nazionale delle Ricerche, Segrate, Italy, 2Radiology, Northwestern University, Chicago, IL, United States
Voxel-by-voxel fitting of intravoxel incoherent motion (IVIM) MRI data using a bi-exponential model is challenging especially with low signal-to-noise ratio (SNR) diffusion-MR images. We propose to combine and use a supervised Deep Neural Network (DNN) approach to increase SNR of the acquired images and thus improve extraction of reliable parameter estimation using a segmented least square fitting algorithm. The effectiveness of the proposed method was demonstrated in both simulated and acquired in vivo data. The proposed approach is promising and can increase performance of the fitting algorithms especially for the case of images with high background noise.
|
||
2526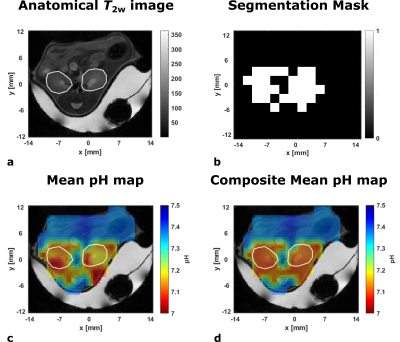 |
58 | Improved detection of multiple kidney pH compartments by deep learning in MRS and MRSI with hyperpolarized 13C-labelled zymonic acid
Martin Grashei1, Wai-Yan Ryana Fok2, Jason G. Skinner1, Bjoern H. Menze2, and Franz Schilling1
1Department of Nuclear Medicine, TUM School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany, 2Department of Informatics, Technical University of Munich, Munich, Germany Accurate determination of peak position is challenging for spectra with dense spectral regions paired with low SNR as occuring in pH measurements using hyperpolarized [1,5-13C2,3,6,6,6-D4]zymonic acid in kidney of mice. Despite scarcity of available data from preclinical experiments, convolutional neural networks (CNN) and multilayer perceptrons (MLP) could be trained by complementing real and augmented data with synthetic spectra. While MLPs do not achieve suitable performance, CNNs predict pH compartments with an accuracy comparable or superior to supervised line fitting in synthetic test spectra. Further, CNNs allow generation of composite pH maps with improved quality while quantitatively agreeing with line-fitted maps. |
||
2527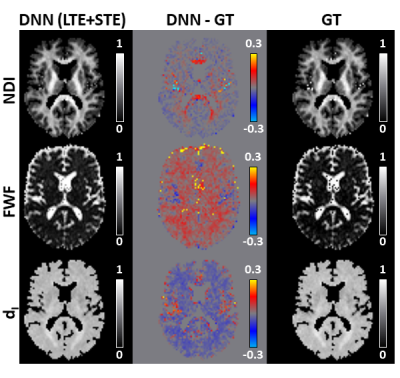 |
59 | Deep learning framework for accelerating revised-NODDI parameter estimation with tensor-valued diffusion encoding
Michele Guerreri1,2, Hojjat Azadbakht2, and Hui Zhang1
1Computer Science & Centre for Medical Image Computing, University College London, London, United Kingdom, 2AINOSTICS, Manchester, United Kingdom This work demonstrates the feasibility of using deep learning (DL) to accelerate revised-NODDI parameter estimation with data acquired using tensor-valued diffusion encoding (TVDE). Revised-NODDI is a recently proposed version of NODDI which showed improved compatibility with TVDE. Thanks to this compatibility the model has an extra free parameter to be estimated which, with conventional fitting methods, further slowdown NODDI’s time-demanding parameter estimation. DL methods can vastly accelerate this process. We show that accurate estimation of revised-NODDI parameters can be obtained via a DL framework. We compare the results with those obtained with conventional fitting methods. |
||
2528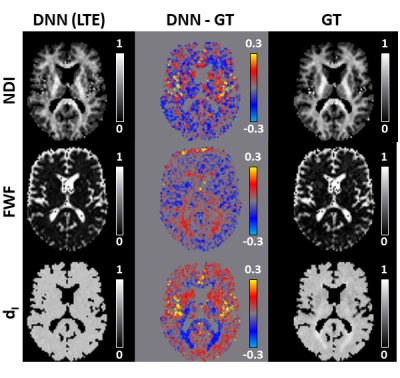 |
60 | Revised-NODDI with conventional dMRI data enabled by deep learning
Michele Guerreri1,2, Hojjat Azadbakht2, and Hui Zhang1
1Computer Science & Centre for Medical Image Computing, University College London, London, United Kingdom, 2AINOSTICS, Manchester, United Kingdom This work shows that deep learning (DL) enables revised-NODDI parameter estimation from conventional dMRI data. Revised-NODDI is a recently proposed model which overcomes some limitations of NODDI. With conventional fitting methods, revised-NODDI parameters can be robustly estimated only in the presence of data acquired with multiple tensor-valued diffusion encodings. However, this new generation of acquisitions is not yet routinely available in clinical research. We show that revised-NODDI parameters estimated using conventional dMRI data via a DL framework are comparable with the parameters estimated fitting the model to data acquired using multiple tensor-valued diffusion encoding. |
||
2529 |
61 | DCE-DRONE: Perfusion MRI Parameter Estimation using a DRONE Neural Network
Soudabeh Kargar1, Ouri Cohen2, and Ricardo Otazo2
1MSKCC, NEW YORK, NY, United States, 2MSKCC, New York, NY, United States
In this work, we demonstrate the estimation of DCE acquired perfusion parameters using a DRONE neural network trained on numeric simulated data. Experiments in a digital phantom are used to demonstrate the feasibility of our approach and the clinical utility is shown in subjects with gynecological tumors.
|
||
2530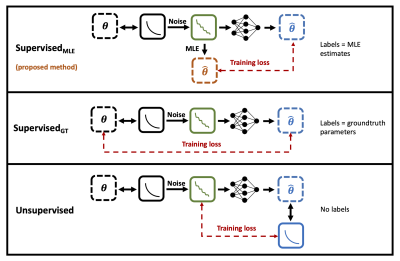 |
62 | Quantitative MRI parameter estimation with supervised deep learning: MLE-derived labels outperform groundtruth labels
Sean Epstein1, Timothy J.P. Bray2, Margaret A. Hall-Craggs2, and Hui Zhang1
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2University College London, London, United Kingdom We propose a novel deep learning technique for quantitative MRI parameter estimation. Our method is trained to map noisy qMRI signals to conventional best-fit parameter labels, which act as proxies for the groundtruth parameters we wish to estimate. We show that this training leads to more accurate predictions of groundtruth model parameters than traditional approaches which train on these groundtruths directly. Furthermore, we show that our proposed method is both conceptually and empirically equivalent to existing unsupervisedapproaches, with the advantage of being formulated as supervised training. |
||
2531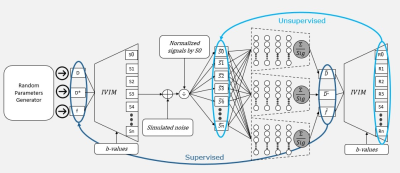 |
63 | SUPER-IVIM-DC, A supervised deep-learning with data consistency approach for IVIM model parameter estimation from Diffusion-Weighted MRI data
Elad Rotman1, Onur Afacan2, Sila Kurugol2, Simon K Warfield2, and Moti Freiman1
1Faculty of Biomedical Engineering, Technion-IIT, Haifa, Israel, 2Computational Radiology lab, Boston Children’s Hospital, Harvard Medical School, Boston, MA, United States
Recently, unsupervised deep-learning showed improved performance in estimating the “Intra-Voxel incoherent motion” (IVIM) signal decay model parameters from Diffusion-weighted Magnetic Resonance Imaging (DW-MRI) data compared to classical methods. However, such deep-learning models do not generalize well on acquisitions with a high signal to noise ratio (SNR). In this work, we introduce SUPER-IVIM-DC, a supervised deep learning network coupled with a data consistency term to improve the capacity of deep-learning-based models to generalize the IVIM signal decay model. We demonstrated an improvement in model generalization, accuracy, and homogeneity using simulation, phantom, and in-vivo experiments.
|
||
2532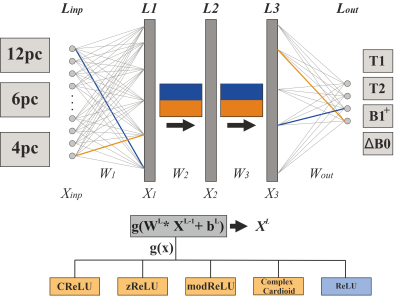 |
64 | Investigating complex-valued neural networks applied to phase-cycled bSSFP for multi-parametric quantitative tissue characterization
Florian Birk1, Julius Steiglechner1, Klaus Scheffler1,2, and Rahel Heule1,2
1Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 2Department of Biomedical Magnetic Resonance, University of Tübingen, Tübingen, Germany
The bSSFP sequence is highly sensitive to relaxation parameters, tissue microstructure, and off-resonance frequencies, which has recently been shown to enable multi-parametric tissue characterization in the human brain using real-valued NNs. In this work, a new approach based on complex-valued NNs for voxel-wise simultaneous multi-parametric quantitative mapping with phase-cycled bSSFP input data is presented, possibly facilitating data handling. Relaxometry parameters (T1, T2) and field map estimates (B1+, ΔB0) could be quantified with high robustness and accuracy. The quantitative results were compared for different activation functions, favoring phase-sensitive implementations.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.