Digital Poster
Machine Learning/Artificial Intelligence: Clinical Application II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1511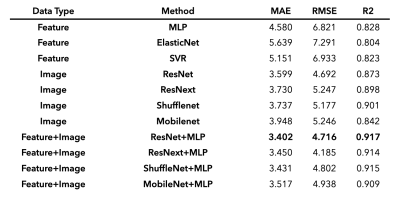 |
61 | Brain age prediction using fusion deep learning combining pre-engineered and convolution-derived features
HeeJoo Lim1,2, Eunji Ha3, Suji Lee3, Sujung Yoon3,4, In Kyoon Lyoo3,4,5, and Taehoon Shin1,2
1Division of Mechanical and Biomedical Engineering, Ewha Womans University, Seoul, Korea, Republic of, 2Graduate Program in Smart Factory, Ewha Womans University, Seoul, Korea, Republic of, 3Ewha Brain Institute, Ewha Womans University, Seoul, Korea, Republic of, 4Department of Brain and Cognitive Sciences, Ewha Womans University, Seoul, Korea, Republic of, 5Graduate School of Pharmaceutical Sciences, Ewha Womans University, Seoul, Korea, Republic of
Prediction of biological brain age is important as its deviation from chronological age can serve as a biomarker for degenerative neurological disorders. In this study, we suggest novel fusion deep learning algorithms which combine pre-engineered features and convolutional neural net (CNN) extracted features of T1-weighted MR images. Over all backbone CNN architectures, fusion models improved prediction accuracy (mean absolute error (MAE) = 3.40–3.52) compared with feature-engineered regression (MAE = 4.58–5.15) and image-based CNN (MAE = 3.60–3.95) alone. These results indicate that using both features derived from convolution and pre-engineering can complement each other in predicting brain age.
|
||
1512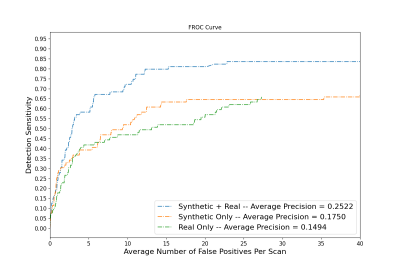 |
62 | Microbleed detection in autopsied brains from community-based older adults using microbleed synthesis and deep learning
Grant Nikseresht1, Ashish A. Tamhane2, Carles Javierre-Petit3, Arnold M. Evia2, David A. Bennett2, Julie A. Schneider2, Gady Agam1, and Konstantinos Arfanakis2,3
1Department of Computer Science, Illinois Institute of Technology, Chicago, IL, United States, 2Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, IL, United States, 3Department of Biomedical Engineering, Illinois Institute of Technology, Chicago, IL, United States
Automated cerebral microbleed (CMB) detection on ex-vivo MRI is key to enabling MRI-pathology studies in large community-based cohorts where manual CMB annotation is time consuming and prone to error. The aim of this study is to develop a CMB detection algorithm to aid in the quantization and localization of CMBs on ex-vivo T2*-weighted gradient echo MRI in community-based cohorts. A CMB synthesis algorithm is proposed and synthetic CMBs are used to train a neural network for CMB detection. A model trained with both synthetic and real data is shown to outperform models trained on synthetic or real data alone.
|
||
1513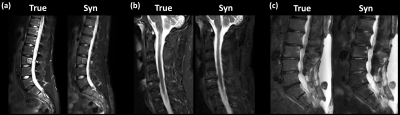 |
63 | Synthetic T2-weighted fat sat delivers valuable information on spine pathologies: multicenter validation of a Generative Adversarial Network
Sarah Schlaeger1, Katharina Drummer1, Malek El Husseini1, Florian Kofler1,2,3, Nico Sollmann1,4,5, Severin Schramm1, Claus Zimmer1, Dimitrios C. Karampinos6, Benedikt Wiestler1, and Jan S. Kirschke1
1Department of Diagnostic and Interventional Neuroradiology, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany, 2Department of Informatics, Technical University of Munich, Munich, Germany, 3TranslaTUM - Central Insitute for Translational Cancer Research, Technical University of Munich, Munich, Germany, 4TUM-NeuroImaging Center, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany, 5Department of Diagnostic and Interventional Radiology, University Hospital Ulm, Ulm, Germany, 6Department of Diagnostic and Interventional Radiology, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany
Generative Adversarial Networks (GANs) can synthesize missing Magnetic Resonance (MR) contrasts from existing MR data. In spine imaging, sagittal T2-w fat sat (fs) sequences are an important additional MR contrast next to conventional T1-w and T2-w sequences. In this study, the diagnostic performance of a GAN-based, synthetic T2-w fs is evaluated in a multicenter dataset. By comparing the synthetic T2-w fs with its true counterpart regarding ability to detect spinal pathologies not seen on T1-w and non-fs T2-w, diagnostics agreement, and image and fs quality our work shows that a synthetic T2-w fs delivers valuable information on spine pathologies.
|
||
1514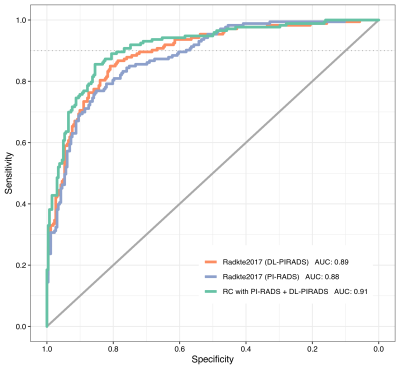 |
64 | Prostate Cancer Risk Assessment using Fully Automatic Deep Learning in MRI: Integration with Clinical Data using Logistic Regression Models
Adrian Schrader1,2, Nils Bastian Netzer1,2, Magdalena Görtz3, Constantin Schwab4, Markus Hohenfellner3, Heinz-Peter Schlemmer1, and David Bonekamp1
1Division of Radiology, German Cancer Research Center, Heidelberg, Germany, 2Heidelberg University Medical School, Heidelberg, Germany, 3Department of Urology, University of Heidelberg Medical Center, Heidelberg, Germany, 4Department of Pathology, University of Heidelberg Medical Center, Heidelberg, Germany For patients with clinical suspicion for significant prostate cancer, the decision to undergo prostate biopsy can be supported by calculating the individual risk profile using demographic and clinical information along with multiparametric MRI assessment. We could show that the prediction performance of an established risk calculator remained stable after substituting manual PI-RADS scores for assessments from a fully automated deep learning system. Combining deep learning and PI-RADS resulted in significant improvements over using only PI-RADS. Complementary information that deep learning models are able to extract enable synergies with radiologists to improve individual risk predictions. |
||
1515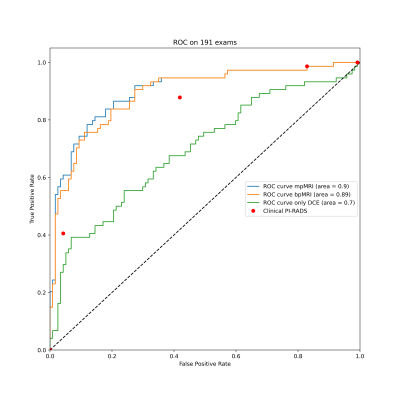 |
65 | Multi-parametric versus bi-parametric prostate MRI for deep learning: Marginal benefits from adding dynamic contrast-enhanced images
Nils Bastian Netzer1,2, Adrian Schrader1,2, Magdalena Görtz3, Constantin Schwab4, Markus Hohenfellner3, Heinz-Peter Schlemmer1, and David Bonekamp1
1Radiology, German Cancer Research Center, Heidelberg, Germany, 2Heidelberg University Medical School, Heidelberg, Germany, 3Urology, University of Heidelberg Medical Center, Heidelberg, Germany, 4Pathology, University of Heidelberg Medical Center, Heidelberg, Germany The value of dynamic contrast enhanced MRI (DCE) for the diagnosis of prostate cancer is unclear and has not yet been investigated in the context of deep learning. We trained 3D U-Nets to segment prostate cancer on bi-parametric MRI and on DCE images of 761 exams. On a test set of 191 exams, the bi-parametric baseline achieved a ROC AUC of 0.89, showing a higher specificity that clinical PI-RADS at a sensitivity of 0.9. Additional improvement could be achieved by fusing bpMRI and DCE predictions, resulting in a ROC AUC of 0.9. |
||
1516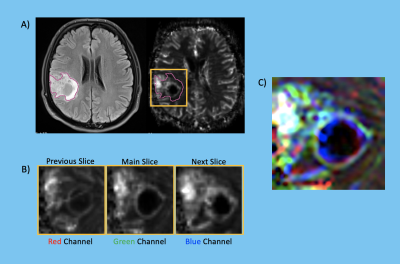 |
66 | Differentiation of IDH and TERTp mutations in Glioma Using Dynamic Susceptibility Contrast MRI with Machine Learning at 3T
Buse Buz-Yalug1, Ayca Ersen Danyeli2,3, Cengiz Yakicier3,4, Necmettin Pamir3,5, Alp Dincer3,6, Koray Ozduman3,7, and Esin Ozturk-Isik1
1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Medical Pathology, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 3Center for Neuroradiological Applications and Reseach, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 4Department of Molecular Biology and Genetics, Istanbul, Turkey, 5Department of Neurosurgery, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 6Department of Radiology, Acıbadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 7Department of Neurosurgery, Acıbadem Mehmet Ali Aydinlar University, Istanbul, Turkey
Isocitrate dehydrogenase (IDH) and telomerase reverse transcriptase promoter (TERTp) mutations highly affect the clinical outcome in gliomas. The aim of this study was to identify IDH and TERTp mutations in glioma patients using machine learning approaches on relative cerebral blood volume (rCBV) maps obtained from dynamic susceptibility contrast MRI (DSC-MRI). The highest classification accuracy was 87.2% (sensitivity = 85.7%, specificity = 88.9%) for the IDH subgroup, 81.8% accuracy (sensitivity = 77.5%, specificity = 86.4%) was obtained for classifying the TERTp subgroup. Additionally, a classification accuracy of 89.6% (sensitivity = 88.3%, specificity = 91.2%) was obtained for identifying the TERTp-only gliomas.
|
||
1517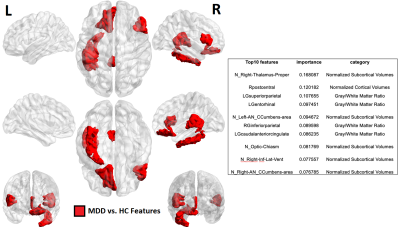 |
67 | Machine learning based classification of major depressive disorder using clinical symptom scales and ultrahigh field MRI features Video Not Available
Gaurav Verma1, Xin Xing2, Yael Jacob3, Bradley N Delman4, James Murrough3, Ai-Ling Lin5, and Priti Balchandani1
1Biomedical Engineering and Imaging Institute, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 2Computer Science, University of Kentucky, Lexington, KY, United States, 3Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 4Diagnostic, Molecular and Interventional Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 5Radiology, University of Missouri, Columbia, MO, United States
Major depression is highly-prevalent disorder with frustratingly-high rates of treatment resistance. Ultrahigh field imaging may provide objective quantitative biomarkers for characterizing depression, generating insight into clinical phenotypes of this heterogeneous disease. Forty-two major depressive disorder patients currently off anti-depressant treatment were recruited for scanning at ultrahigh field, and given batteries of clinical symptom measures. Machine-learning clustering analysis was performed to group patients by clinical symptoms and differences in imaging features observed. A separate analysis was performed in the reverse direction clustering on quantified imaging features and identifying clinical differences between clusters, including differences in ruminative response between the patient clusters.
|
||
1518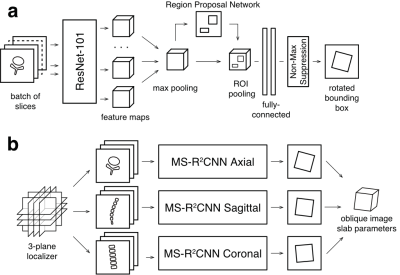 |
68 | Towards Clinical Translation of Machine Learning-based Automated Prescription of Spine MRI Acquisitions
Eugene Ozhinsky1, Felix Liu1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States
High quality scan prescription that optimally covers the area of interest with scan planes aligned to relevant anatomical structures is crucial for error-free radiologic interpretation. In this study we used images and metadata from previously acquired examinations of lumbar spine to train machine learning-based automated prescription models without the need of any manual annotation or feature engineering. The automated prescription pipeline was integrated with the scanner console software and evaluated in healthy volunteer experiments. This study demonstrates the feasibility of using oriented object detection-based pipelines on the scanner for automated prescription of lumbar spine acquisitions.
|
||
1519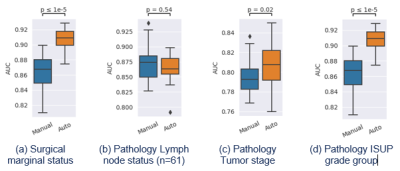 |
69 | Comparison of whole-prostate radiomics models of disease severity derived from expert and AI based prostate segmentations
Paul E Summers1, Lars Johannes Isaksson2, Matteo Johannes Pepa2, Mattia Zaffaroni2, Maria Giulia Vincini2, Giulia Corrao2,3, Giovanni Carlo Mazzola2,3, Marco Rotondi2,3, Sara Raimondi4, Sara Gandini4, Stefania Volpe2,3, Zaharudin Haron5, Sarah Alessi1, Paola Pricolo1, Francesco Alessandro Mistretta6, Stefano Luzzago6, Federico Cattani7, Gennaro Musi3,6, Ottavio De Cobelli3,6, Marta Cremonesi8,
Roberto Orecchia9, Giulia Marvaso2,3, Barbara Alicja Jereczek-Fossa2,3, and Giuseppe Petralia3,10
1Division of Radiology, IEO, European Institute of Oncology IRCCS, Milano, Italy, 2Division of Radiation Oncology, IEO, European Institute of Oncology IRCCS, Milano, Italy, 3Department of Oncology and Hemato-oncology, University of Milan, Milano, Italy, 4Department of Experimental Oncology, IEO, European Institute of Oncology IRCCS, Milano, Italy, 5Radiology Department, National Cancer Institute, Putrajaya, Malaysia, 6Division of Urology, IEO, European Institute of Oncology IRCCS, Milano, Italy, 7Unit of Medical Physics, IEO, European Institute of Oncology IRCCS, Milano, Italy, 8Radiation Research Unit, IEO, European Institute of Oncology IRCCS, Milano, Italy, 9Scientific Directorate, IEO, European Institute of Oncology IRCCS, Milano, Italy, 10Precision Imaging and Research Unit, IEO, European Institute of Oncology IRCCS, Milano, Italy
A persisting concern is that downstream models of clinical endpoints may depend on whether the contours were drawn by an expert or an AI. Prediction models for surgical margin status, and pathology-based lymph nodes, tumor stage and ISUP grade group were formed using clinical and radiological features along with whole-prostate radiomic features based on manual and AI segmentations of the prostate in 100 patients who proceeded to prostatectomy after multiparametric-MRI. The models based on AI segmented prostates differed from those based on manual segmentation, but with similar if not better performance. Further testing of generalizability of the models is required.
|
||
1520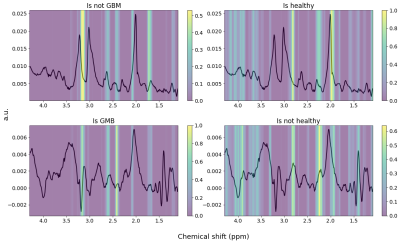 |
70 | A Deep Neural Network for Detection of Glioblastomas in Spectroscopic MRI
Erin Beate Bjørkeli1,2, Jonn Terje Geitung1,2, and Morteza Esmaeili1,3
1Department of Diagnostic Imaging, Akershus University Hospital, Oslo, Norway, 2Institue of Clinical Medicine, University of Oslo, Oslo, Norway, 3Department of Electrical Engineering and Computer Science, University of Stavanger, Stavanger, Norway
We have developed a MRSI spectra classifying convolutional neural network (CNN), building on the AUTOMAP model for image and spectra reconstruction. The model was trained to discern between non-water-suppressed spectra from healthy subjects and glioblastoma (GBM) patients. The trained CNN was able to classify the spectra correctly and seemed to recognize the healthy spectra based on the NAA-peak and the GBM based on the choline levels and possibly 2HG, indicating an IDH mutation.
|
||
1521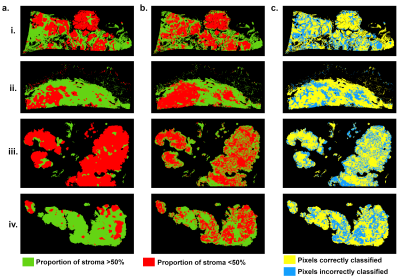 |
71 | Predicted tumor stroma segmentation from high-field MR texture maps and machine learning: an ex vivo study on ovarian tumors
Marion Tardieu1, Lakhdar Khellaf2, Maida Cardoso3, Olivia Sgarbura4, Pierre-Emmanuel Colombo4, Christophe Goze-Bac3, and Stephanie Nougaret1
1Montpellier Cancer Research Institute (IRCM), INSERM U1194, University of Montpellier, Montpellier, France, 2Department of pathology, Montpellier Cancer Institute (ICM), Montpellier, France, 3BNIF facility, L2C, UMR 5221, CNRS, University of Montpellier, Montpellier, France, 4Department of Surgery, Montpellier Cancer Institute (ICM), Montpellier, France
The objective was to probe the associations of high-field MR-images and their derived texture maps (TM) with histopathology in ovarian cancer (OC). Four ovarian tumors were imaged ex-vivo using a 9.4T-MR scanner. Automated MR-derived stroma-tumor segmentation maps were constructed using machine learning and validated against histology. Through TM, we found that areas of tumor cells appeared uniform on MR-images, while areas of stroma appeared heterogeneous. Using the automated model, MRI predicted stromal proportion with an accuracy from 61.4% to 71.9%. In this hypothesis-generating study, we showed that it is feasible to resolve histologic structures in OC using ex-vivo MR radiomics.
|
||
1522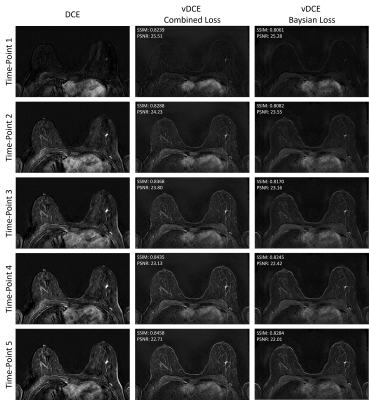 |
72 | Virtual Dynamic Contrast Enhanced MRI of the Breast using a U-Net
Hannes Schreiter1,2, Vishal Sukumar1,2, Lorenz Kapsner1, Lukas Folle2, Sabine Ohlmeyer1, Frederik Bernd Laun1, Evelyn Wenkel1, Michael Uder1, Andreas Maier2, Sebastian Bickelhaupt1, and Andrzej Liebert1
1Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nürnberg (FAU), Erlangen, Germany, 2Patter Recognition Lab, Department of Computer Science, Friedrich-Alexander University Erlangen-Nürnberg (FAU), Erlangen, Germany Magnetic resonance imaging (MRI) examinations of the breast require intravenous administration of gadolinium based contrast agents (GBCA) for comprehensive characterization of the tissue. Novel approaches reducing the need for GBCA might therefore be of value. Here a virtual dynamic contrast enhancement (vDCE) using a U-net architecture is investigated in a cohort of n=540 patients. The vDCE generates T1 subtraction images for five consecutive time points predicting the perfusion maps based on native T1-weighted, T2-weighted, and multi-b-value diffusion weighted acquisitions. A mean structural similarity index (SSIM) value over a test group of 82 patients of 0.848±0.025 was achieved. |
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.