Digital Poster
Imaging Processing & Analysis
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1961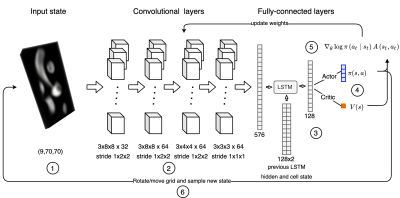 |
56 | Automatic 3D to 2D reformatting in 4D flow MRI using continuous reinforced learning
Javier Bisbal1,2,3, Julio Sotelo1,3,4, Cristobal Arrieta1,3, Pablo Irarrázabal1,2,3, Marcelo Andia1,3,5, Cristian Tejos1,2,3, and Sergio Uribe1,3,5
1Biomedical Imaging Center UC, Pontificia Universidad Catolica de Chile, Santiago, Chile, 2Electrical Engineering Department, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 3ANID – Millennium Science Initiative Program – Millennium Nucleus for Cardiovascular Magnetic Resonance, Santiago, Chile, 4School of Biomedical Engineering, Universidad de Valparaíso, Valparaíso, Chile, 5Department of Radiology, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile
One major limitation on 4D flow MRI is the time-consuming and user-dependent post-processing. We developed an automated reinforced deep learning framework for plane planning in 4D flow data. This method sequentially updates plane parameters towards a target plane based on a continuous policy. A total of 83 4D flow MRI scans were considered, 41 for training, 14 for validation and 28 for test. Our method achieves good results in terms of angulation and distance error (9.21 ± 3.85 degrees and 3.72 ± 2.19 mm).
|
||
1962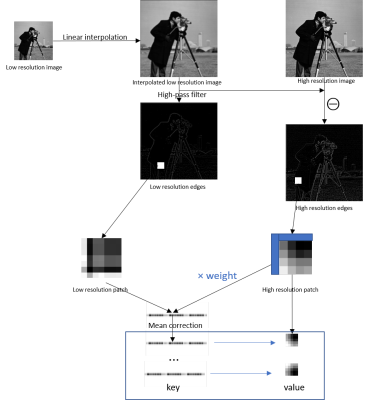 |
57 | Single-image superresolution of hyperpolarized 13C spectroscopic images
Kofi Deh1, Nathaniel Kim1, Guannan Zhang1, Miloushev Vesselin1, and Kayvan Keshari1
1Memorial Sloan Kettering Cancer Center, New York, NY, United States
Hyperpolarized 13C spectroscopic images are acquired at a low spatial resolution, making it necessary to apply superresolution techniques to the metabolite maps prior to fusion with the proton anatomic image for visualization of metabolite biodistribution. In this work, we demonstrate great improvement in image quality for preclinical and clinical images when the metabolite map is upsampled by high spatial frequency transfer from magnetic resonance images of thermally polarized 13C phantoms to the invivo metabolic image.
|
||
1963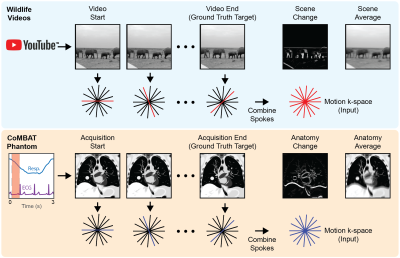 |
58 | Learning motion correction from YouTube for real-time MRI reconstruction with AUTOMAP
David E J Waddington1, Christopher Chiu1, Nicholas Hindley1,2, Neha Koonjoo2, Tess Reynolds1, Paul Liu1, Bo Zhu2, Chiara Paganelli3, Matthew S Rosen2,4,5, and Paul J Keall1
1ACRF Image X Institute, Faculty of Medicine and Health, The University of Sydney, Sydney, Australia, 2A. A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, Milan, Italy, 4Department of Physics, Harvard University, Cambridge, MA, United States, 5Harvard Medical School, Boston, MA, United States
Today’s MRI does not have the spatio-temporal resolution to image the anatomy of a patient in real-time. Therefore, novel solutions are required in MRI-guided radiotherapy to enable real-time adaptation of the treatment beam to optimally target the cancer and spare surrounding healthy tissue. Neural networks could solve this problem, however, there is a dearth of sufficiently large training data required to accurately model patient motion. Here, we use the YouTube-8M database to train the AUTOMAP network. We use a virtual dynamic lung tumour phantom to show that the generalized motion properties learned from YouTube lead to improved target tracking accuracy.
|
||
1964 |
59 | MR-Class: MR Image Classification using one-vs-all Deep Convolutional Neural Network
Patrick Salome1,2,3,4, Francesco Sforazzini1,2,3,4, Andreas Kudak3,5,6, Matthias Dostal3,5,6, Nina Bougatf3,5,6, Jürgen Debus3,4,5,7, Amir Abdollahi1,3,4,5, and Maximilian Knoll1,3,4,5
1CCU Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Medical Faculty, Heidelberg University Hospital, Heidelberg, Germany, 3Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg, Germany, 4German Cancer Consortium (DKTK) Core Center, Heidelberg, Germany, 5Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany, 6CCU Radiation Therapy, German Cancer Research Center (DKFZ), Heidelberg, Germany, 7National Center for Tumor Diseases (NCT), Heidelberg, Germany MR-Class is a deep learning-based MR image classification tool that facilitates and speeds up the initialization of big data MR-based research studies by providing fast, robust, and quality-assured MR image classifications. It was observed in this study that corrupt and misleading DICOM metadata could lead to a misclassification of about 10%. Therefore, in a field where independent datasets are frequently needed for study validations, MR-Class can eliminate the cumbrousness of data cohorts curation and sorting. This can greatly impact researchers interested in big data multiparametric MRI studies and thus contribute to the faster deployment of clinical artificial intelligence applications. |
||
1965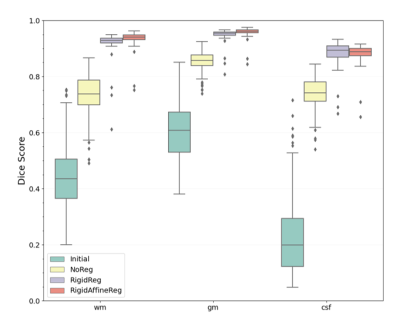 |
60 | Medical Image Registration Using Deep Learning Techniques Applied to Pediatric Magnetic Resonance Imaging (MRI) Brain Scans
Andjela Dimitrijevic1,2, Vincent Noblet3, and Benjamin De Leener1,2,4
1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montréal, Montréal, QC, Canada, 2Research Center, Ste-Justine Hospital University Centre, Montréal, QC, Canada, 3ICube-UMR 7357, Université de Strasbourg, CNRS, Strasbourg, France, 4Computer Engineering and Software Engineering, Polytechnique Montréal, Montréal, QC, Canada
Deep learning techniques have a potential in allowing fast deformable registration tasks. Studies around registration often focus on adult populations, even if there is a need for pediatric research where less data and studies are being produced. In this study, we compared three methods for intra-subject registration on publicly available Calgary Preschool dataset. Using the DeepReg framework, pre-registering with a rigid and affine transformation (proposed RigidAffineReg method) showed the least negative JD values and the highest Dice score (0.924±0.045). By achieving faster alignments, this tool for pediatric MRI scans could help proliferate larger scale population research in brain developmental studies.
|
||
1966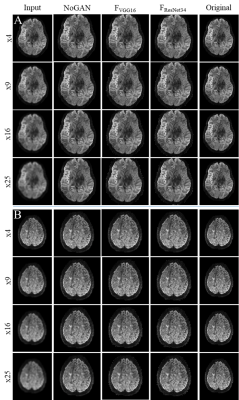 |
61 | Deep Learning with Perceptual Loss Enables Super-Resolution in 7T Diffusion Images of the Human Brain
David Lohr1, Theresa Reiter1,2, and Laura Schreiber1
1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), Würzburg, Germany, 2Department of Internal Medicine I, University Hospital Würzburg, Würzburg, Germany
Diffusion weighted imaging has become a key imaging modality for the assessment of brain connectivity as well as the structural integrity, but the method is limited by low SNR and long scan times. In this study we demonstrate that AI models trained on a moderate number of publicly available 7T datasets (n=12) are able to enhance image resolutions in diffusion MRI up to 25 times. The applied NoGAN model performs well for smaller resolution enhancements (4- and 9-fold), but generates "hyper" realistic images for higher enhancements (16- and 25-fold). Models trained using perceptual loss seem to avoid this limitation.
|
||
1967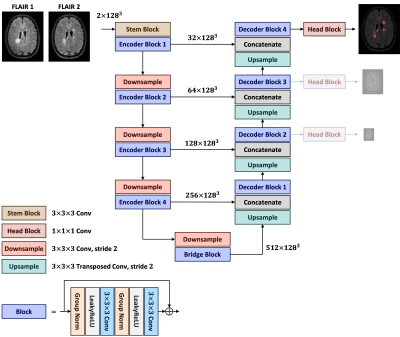 |
62 | Longitudinal Multiple Sclerosis Lesion Segmentation Using Pre-activation U-Net
Pooya Ashtari1,2, Berardino Barile1,2, Dominique Sappey-Marinier2, and Sabine Van Huffel1
1Department of Electrical Engineering (ESAT), KU Leuven, Leuven, Belgium, 2CREATIS (CNRS UMR5220 & INSERM U1294), Université Claude-Bernard Lyon 1, Lyon, France
Automated segmentation of new multiple sclerosis (MS) lesions in MRI data is crucial for monitoring and quantifying MS progression. Manual delineation of such lesions is laborious and time-consuming since experts need to deal with 3D images and numerous small lesions. We propose a 3D encoder-decoder architecture with pre-activation blocks to segment new MS lesions in longitudinal FLAIR images. We also applied intensive data augmentation and deep supervision to mitigate the limited data and the class imbalance problem. The proposed model, called Pre-U-Net, achieved a Dice score of 0.62 and a sensitivity of 0.58 on the public challenge MSSEG-2 dataset.
|
||
1968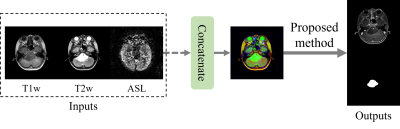 |
63 | A Mask Guided Attention Generative Adversarial Network for Contrast-enhanced T1-weight MR Synthesis
Yajing Zhang1, Xiangyu Xiong1, and Chuanqi Sun1
1MR Clinical Science, Philips Healthcare, Suzhou, China Image synthesis methods based on deep learning has recently achieved success in reducing the dosage of gadolinium-based contrast agents (GBCAs). However, these methods cannot focus on the region of interest to synthesize realistic images. To address this issue, a mask guided attention generative adversarial network (MGA-GAN) was proposed to synthesize contrast enhanced T1-weight images from the multi-channel inputs. Qualitive and quantitative results indicate that the proposed MGA-GAN can improve the synthesized images with higher quality for details of brainstem glioma, compared with state-of-the-art methods. |
||
1969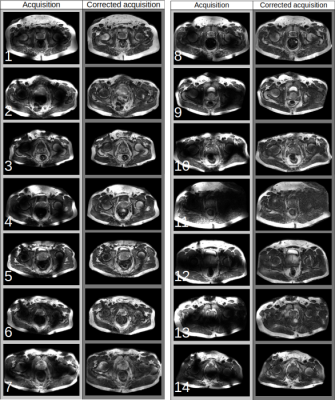 |
64 | Retrospective correction of B1 field inhomogeneities in T2w 7T prostate patient data
Seb Harrevelt1, Daan Reesink2, Astrid Lier, van3, Richard Meijer3, Josien Pluim4, and Alexander Raaijmakers1
1TU Eindhoven, Utrecht, Netherlands, 2Meander Medisch Centrum, Utrecht, Netherlands, 3UMC Utrecht, Utrecht, Netherlands, 4TU Eindhoven, Eindhoven, Netherlands
Prostate imaging at ultra-high fields is heavily affected by B1 field induced inhomogeneities. This not only results in unattractive images but it also might affect clinical diagnosis . To remedy this we developed a deep learning model that retrospectively corrects for the bias field. We applied this model to a clinical data set and demonstrated its performance in a qualitative manner. The results indicate that the model is able to drastically reduce the inhomogeneities in a variety of cases while the tissue contrast is generally maintained and the underlying anatomy has been successfully recovered.
|
||
1970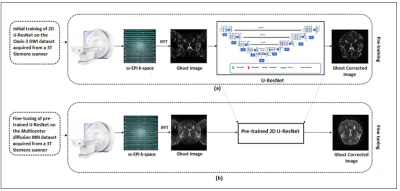 |
65 | EPI Nyquist Ghost Artifact Correction for Brain Diffusion Weighted Imaging (DWI) using Deep Learning
Fatima Sattar1, Sadia Ahsan1, Fariha Aamir1, Ibtisam Aslam1,2, Iram Shahzadi3,4, and Hammad Omer1
1Medical Image Processing Research Group (MIPRG), Department of Electrical and Computer Engineering, COMSATS University, Islamabad, Pakistan, 2Service of Radiology, Geneva University Hospitals and Faculty of Medicine, Hospital University of Geneva and University of Geneva, Geneva, Switzerland, 3OncoRay – National Center for Radiation Research in Oncology, Faculty of Medicine and University Hospital Carl Gustav Carus, Technische Universität Dresden, Helmholtz-Zentrum Dresden –Rossendorf, Dresden, Germany, Dresden, Germany, 4German Cancer Research Center (DKFZ), Heidelberg, Germany, Dresden, Germany
Echo-planar imaging suffers from Nyquist ghost (i.e., N/2 ghost) artifacts because of poor system gradients and delays. Many conventional methods have been used in literature to remove N/2 artifacts in Diffusion Weighted Imaging (DWI) but often produce erroneous results. This paper presents a deep learning approach to eliminate the phase error of k-space for removing the Nyquist ghost artifacts in DWI. Experimental results show successful removal of the ghost artifacts with improved SNR and reconstruction quality with the proposed method.
|
||
1971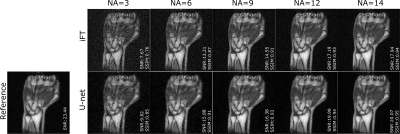 |
66 | Residual U-net for denoising 3D low field MRI
Tobias Senft1, Reina Ayde1, Marco Fiorito1, Najat Salameh1, and Mathieu Sarracanie1
1Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland
Low magnetic field (LF) MRI is currently gaining momentum as a complementary, more flexible and cost-effective approach to MRI diagnosis. However, the impaired signal-to-noise ratio challenges its relevance for clinical applications. Recently, denoising of low SNR images using deep learning techniques has shown promising results for MRI applications. In this study, we assess the denoising performance of residual U-net architecture on different SNR levels of LF MRI data (0.1 T). The model performance has been evaluated on both simulated and acquired LF MRI datasets.
|
||
1972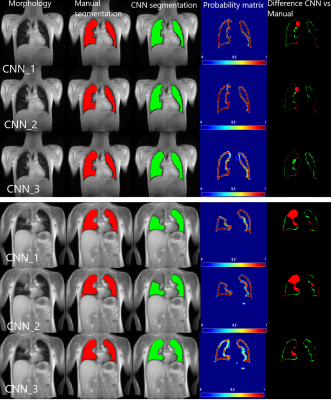 |
67 | Artificially-generated Consolidations and Balanced Augmentation increase Performance of U-Net for Lung Parenchyma Segmentation on MR Images
Cristian Crisosto1,2, Andreas Voskrebenzev1,2, Marcel Gutberlet1,2, Filip Klimeš1,2, Till Kaireit1,2, Gesa Pöhler1,2, Tawfik Alsady1,2, Lea Behrendt1,2, Robin Müller1,2, Maximilian Zubke1,2, Frank Wacker1,2, and Jens Vogel-Claussen1,2
1Institute of Diagnostic and Interventional Radiology, Medical School Hannover, Hannover, Germany, 2Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), Member of the German Centre for Lung Research (DZL), Hannover, Germany, Hannover, Germany
Accurate fully automated lung segmentation is needed to facilitate Fourier-Decomposition employment-based techniques in clinical routine among different centers. However, the lung parenchyma segmentation remains challenging for convolutional neural networks (CNN) when consolidations are present. To improve training balanced augmentation (BA) and artificially-generated consolidations (AGC) were introduced. The proposed CNN was compared to conventional CNNs without BA and AGC using Sørensen-Dice coefficient (SDC) and Hausdorff coefficient (HD). The SDC / HD of the proposed model is significantly higher (p of 0.0001 and p of 0.0146 / p of 0.0009 and p of 0.0152) when compared to CNNs without BA and AGC.
|
||
1973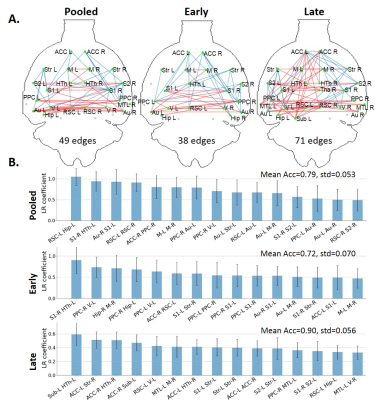 |
68 | Subject classification based on functional connectivity and white matter microstructure in a rat model of Alzheimer’s using machine learning
Yujian Diao1,2,3, Catarina Tristão Pereira2,4, Ting Yin2, and Ileana Ozana Jelescu2,5
1Laboratory of Functional and Metabolic Imaging, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 3Animal Imaging and Technology, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 4Faculdade de Ciências da Universidade de Lisboa, Lisbon, Portugal, 5Department of Radiology, Lausanne University Hospital, Lausanne, Switzerland
Impaired brain glucose consumption is a possible trigger of Alzheimer’s disease (AD). Previous work revealed affected brain structure and function by insulin resistance in terms of functional connectivity and white matter microstructure in a rat model of AD. Here, functional and structural metrics were further used to classify Alzheimer’s from control rats using logistic regression. Our study highlights the MRI-derived biomarkers that best discriminate Alzheimer’s vs control rats early in the course of the disease, with potential translation to human AD.
|
||
1974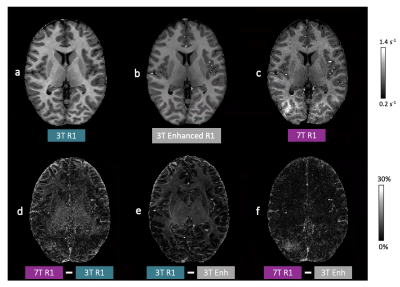 |
69 | Mapping human brain development at new spatial resolutions using deep learning and high-resolution quantitative MRI Video Permission Withheld
Georgia Doumou1,2, Hongxiang Lin2,3, Sara Lorio1, Lenka Vaculčiaková4, Kerrin J. Pine4, Nikolaus Weiskopf4,5, Jonathan O'Muircheartaigh6,7,8, Daniel C. Alexander2, and David W. Carmichael 1
1Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 3Research Center for Healthcare Data Science, Zhejiang Lab, Hangzhou, China, 4Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 5Felix Bloch Institute for Solid State Physics, Faculty of Physics and Earth Sciences, Leipzig University, Leipzig, Germany, 6Department of Forensic & Neurodevelopmental Sciences, King's College London, London, United Kingdom, 7Centre for the Developing Brain, Department of Perinatal Imaging and Health, King's College London, London, United Kingdom, 8MRC Centre for Neurodevelopmental Disorders, King's College London, London, United Kingdom
High-resolution quantitative MRI using ultra-high field scanners (7T) could advance a range of research and clinical applications if limitations, both practical (e.g. long acquisitions) and technical (e.g. B1 non-uniformity), can be avoided. This could be achieved by using 7T information to enhance conventional field strength images. To test this approach, paired 3T-7T R1 maps were used to train a U-Net variant to enhance 3T R1 maps. Leave-one-out cross-validation, quantitative evaluation, as well as external validation on an external clinical dataset, demonstrated promising enhancement with visual and quantitative metrics more similar to 7T R1 maps than the 3T equivalents.
|
||
1975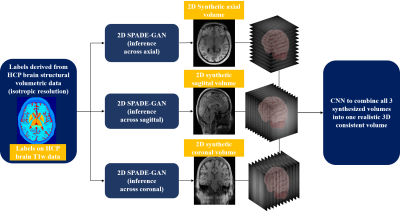 |
70 | 3D brain MRI synthesis utilizing 2D SPADE-GAN and 3D CNN architecture
Aymen Ayaz1, Ronald de Jong1, Samaneh Abbasi-Sureshjani1, Sina Amirrajab1, Cristian Lorenz2, Juergen Weese2, and Marcel Breeuwer1,3
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3MR R&D – Clinical Science, Philips Healthcare, Best, Netherlands
We propose a method to synthesize 3D consistent brain MRI utilizing multiple 2D conditional SPatially-Adaptive (DE)normalization (SPADE) GANs to preserve the spatial information of patient-specific brain anatomy and a 3D CNN based network to improve the image consistency in all directions in a cost-efficient manner. Three individual 2D SPADE-GAN networks are trained across the axial, sagittal and coronal slice directions and their outputs are thereafter used further to train a CNN based 3D image restoration network to combine them into one 3D volume, using real MRI as a reference. The resulting predicted-synthesized 3D brain MRI is evaluated quantitatively and qualitatively.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.