Digital Poster
Relaxation & Multiparameter Mapping II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1336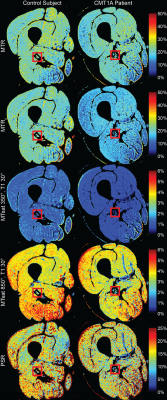 |
42 | Comparison of magnetization transfer quantification methods in the sciatic nerve of Charcot-Marie-Tooth Type 1A patients
Alison Roth1, Yongsheng Chen2, Jun Li2,3, and Richard D. Dortch1,4,5
1Division of Neuroimaging Research, Barrow Neurological Institute, Phoenix, AZ, United States, 2Department of Neurology, Wayne State University, Detroit, MI, United States, 3Department of Neurology, Vanderbilt University, Nashville, TN, United States, 4Vanderbilt University Institute of Imaging Science, Vanderbilt University, Nashville, TN, United States, 5Department of Biomedical Engineering, Vanderbilt University, Nashville, TN, United States
Magnetization transfer (MT) can be quantified as the MT ratio (MTR), MT saturation (MTsat), and quantitative MT (qMT) pool-size-ratio (PSR). MTR is a promising peripheral nerve imaging biomarker in inherited neuropathies but has lower pathological specificity compared to MTsat and PSR. Here, MTR and MTsat metrics are compared to PSR. Eighteen subjects received 16 qMT scans, five T1-weighted scans, and the B0 and B1 fields were measured. Mean MTR, MTsat, and PSR were extracted from the sciatic nerve. MTR had the highest SNR (median>15.76) and scan-rescan repeatability (ICC=0.93, CV=4.6%), whereas MTsat had the strongest correlation to PSR (r=0.94, p<0.001).
|
||
1337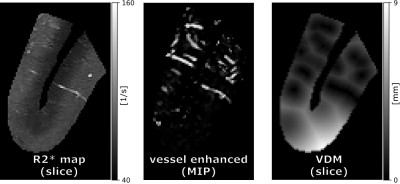 |
43 | Post mortem study of R2* and vessel distance maps across cortical depth
Hendrik Mattern1, Frank Angenstein2, Christian Mawrin3, and Valentina Perosa2,4,5
1Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2German Center for Neurodegenerative Disease, Magdeburg, Germany, 3Institute of Neuropathology, Medical Faculty at the Otto von Guericke University, Magdeburg, Germany, 4J. Philip Kistler Stroke Research Center, Massachusetts General Hospital, Boston, MA, United States, 5Department of Neurology, Otto-von-Guericke University, Magdeburg, Germany
In this proof-of-principle study, an image acquisition and processing pipeline was developed to assess the cortical vasculature and its surrounding tissue. To that end, a block of human brain, which included cortex and juxtacortical white matter, was scanned post-mortem at high resolution with a multi-echo GRE sequence at 9.4T. Subsequently, R2* and vessel distance maps (VDM) were computed and their layer-specific patterns analyzed.
|
||
1338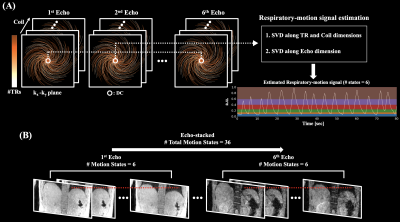 |
44 | Free-Breathing R2* Mapping for Hepatic Iron Quantification Using Respiratory Motion-Resolved 3D Multi-Echo UTE Cones MRI
MungSoo Kang1,2, Yan Wen3, Michael Carl3, Gerald G. Behr4, Ricardo Otazo1,4, and Youngwook Kee1
1Department of Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Department of Biomedical Engineering, Ulsan National Institute of Science and Technology, Ulsan, Korea, Republic of, 3GE Healthcare, Waukesha, WI, United States, 4Department of Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
Respiratory motion is one of the major factors hindering accurate R2* mapping for hepatic iron quantification. In this work, motion-resolved 3D multi-echo UTE cones MRI with pseudo-random view ordering was implemented for motion-robust and free-breathing R2* mapping of hepatic iron. Compared to conventional gridding reconstruction, motion-resolved reconstruction reduced the overestimation of R2* induced by respiratory motion artifacts, enabling an accurate measurement of R2*. 3D multi-echo UTE cones MRI with motion-resolved reconstruction demonstrated the feasibility of accurate free-breathing R2* mapping for hepatic iron quantification.
|
||
1339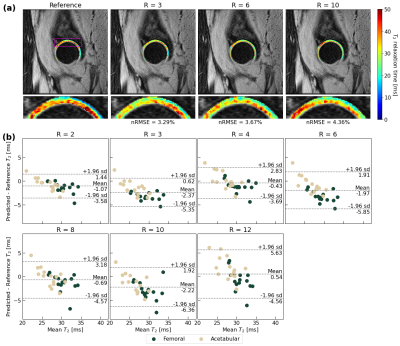 |
45 | A Recurrent Encoder-Decoder Network Accelerates T2 Mapping in Knee, Hip and Lumbar Spine
Aniket Tolpadi1,2, Francesco Calivà1, Misung Han1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Bioengineering, University of California, Berkeley, Berkeley, CA, United States
Compositional MRI (cMRI) offers sensitivity to biochemical changes that can precede morphological changes in tissues and offer quantitative assessments of musculoskeletal tissue health but suffer from long scan times. We present a recurrent encoder-decoder architecture that predicts ground truth T2 maps from spatially undersampled echo images. Reconstructed maps showed strong fidelity to ground truth in lumbar spine IVDs and knee and hip knee cartilage. Reconstruction errors were below clinically significant T2 changes through R=12 in the knee and lumbar spine and R=8 in the hip. This work marks progress towards a clinical pipeline that reduces cMRI acquisition time.
|
||
1340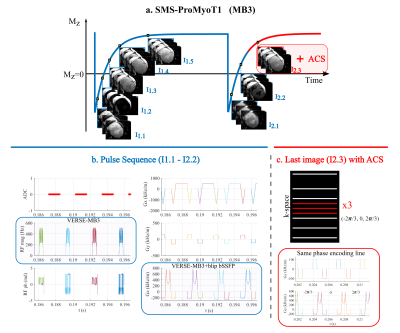 |
46 | Open-Source Myocardial T1 mapping accelerated with SMS: combining an auto-calibrated blip-bSSFP readout with VERSE-MB pulses
Andreia S Gaspar1, Nuno A Silva2, and Rita G Nunes1
1Institute for Systems and Robotics - Lisboa and Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisboa, Portugal, 2Hospital da Luz Learning Health, Luz Saúde, Lisbon, Portugal
Clinical myocardial T1 mapping protocols typically acquire three 2D slices, each obtained in a separate breath-hold; this is only feasible in patients able to do sequential breath-holds. We propose SMS-ProMyoT1 which integrates simultaneous multi-slice (SMS) with open-source ProMyoT1, previously implemented with Pulseq, to obtain all slices in a fast single shot. SMS-ProMyoT1 differs from previous implementations by combining blip-bSSFP with VERSE-multiband for RF modulation, and auto-calibrated blip patterns for a fast self-contained sequence. The method was successfully applied both in a reference phantom and in-vivo.
|
||
1341 |
47 | Feasibility of map recalibration using an in-scan reference system for two MRI mapping cardiac sequences
Davide Cicolari1, Domenico Lizio2, Patrizia Pedrotti3, Monica Teresa Moioli2, Alessandro Lascialfari1, Manuel Mariani1, and Alberto Torresin2,4
1Department of Physics, University of Pavia, Pavia, Italy, 2Department of Medical Physics, ASST Grande Ospedale Metropolitano Niguarda, Milan, Italy, 3Department of Cardiology, ASST Grande Ospedale Metropolitano Niguarda, Milan, Italy, 4Department of Physics, University of Milan, Milan, Italy In this work we tested the feasibility of MRI relaxation time maps recalibration by employing an in-scan reference system. This ‘belt phantom’ was characterized through NMR standard spectroscopic technique. Scan-dependent recalibrations of the relaxation time maps could be performed relying on the ground-truth NMR values of the phantom, aiming to clinical intra- and inter-center harmonization. The in-scan reference phantom allowed also, together with the analysis of the standard deviation maps (measured as the 68% confidence bound of the fitted relaxation time value), to evaluate the reliability of the maps and the applicability of the recalibration. |
||
1342 |
48 | Validation of fast in vivo T1 and T2* mapping of tissues in the knee using 3D UTE
Maik Rothe1, Andreas Deistung1, Richard Brill1, Walter Alexander Wohlgemuth1, and Alexander Gussew1
1University clinic and policlinic for Radiology, University Hospital Halle (Saale), Halle (Saale), Germany In this study we present a 3D high resolution method for fast in vivo T1 and T2* mapping of fast relaxating tissues of the knee. All experiments were performed using a prototypical 3D spoiled gradient echo ultrashort echo time sequence with stack of spirals readout. T1 and T2* values of the mapping technique were validated by reference methods and literature values and showed good agreement for fast T1 and T2* values. The mapping technique enables T1 and T2* quantitation of fast relaxating tissues in a clinically appropriate measurement time of 9 minutes with a submillimeter spatial resolution (0.8mmx0.8mmx0.8mm). |
||
1343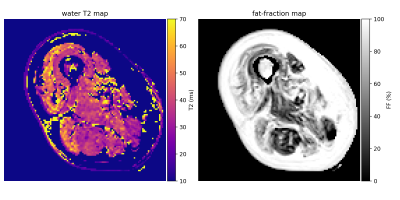 |
49 | A hybrid EPG-hGLLiM method for accurate fat-signal modeling in skeletal muscle T2 mapping
Pierre-Yves Baudin1,2, Ericky Caldas de Almeida Araujo1,2, Harmen Reyngoudt1,2, and Benjamin Marty1,2
1NMR Laboratory, Institute of Myology, Neuromuscular Investigation Center, Paris, France, 2NMR Laboratory, CEA, DRF, IBFJ, MIRCen, Paris, France
In order to improve the reliability of the muscle T2 as a clinical outcome in clinical studies of neuromuscular disorders, we investigate the accuracy of the fat signal modeling in transverse relaxometry from multi-spin echo imaging. A new approach for muscle T2/fat-fraction mapping is proposed, combining a water signal dictionary with water T2/B1-dependent entries created from EPG simulations, and a hybrid Gaussian Locally Linear Mapping (hGLLiM) to provide B1-dependent fat signals. Preliminary results on a dataset of healthy controls, DMD and IBM patients are consistent with common knowledge, but interesting divergences are found when compared to another recent approach.
|
||
1344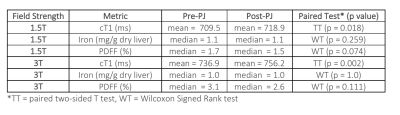 |
50 | Measuring the Impact of Pineapple Juice on Quantitative Liver MRI Metrics cT1, Iron and PDFF at 1.5T and 3T Video Permission Withheld
Yi-Chun Wang1,2, Faezeh Sanaei-Nezhad1, Joao Peixoto1, Carolina Fernandes1, Alex Smith1, Matthew Robson1, and Rajarshi Banerjee1
1Perspectum Ltd., Oxford, United Kingdom, 2University of Oxford, Oxford, United Kingdom
Pineapple juice (PJ) is used in Magnetic Resonance Cholangiopancreatography to suppress gastrointestinal tract signals. To explore how PJ affects multiparametric liver MRI, 30 participants underwent scans before and after ingesting PJ. Image data were analysed with LiverMultiScan (yielding iron corrected T1 (cT1), iron, and PDFF) and statistically compared. The changes post PJ administration were statistically significant for cT1 at both 1.5T and 3T but not for iron and PDFF. However, the repeatability analysis indicates the post PJ administration cT1 changes were smaller than the repeatability limits of agreement, suggesting that PJ has neglect-able clinical effect on multiparametric liver MRI.
|
||
1345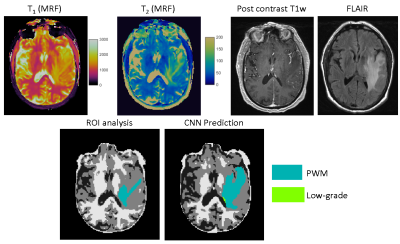 |
51 | Improving annotation accuracy in MRI data using MR Fingerprinting and deep learning
Yong Chen1, Rasim Boyacioglu1, Gamage Sugandima Nishadi Weragoda2, Michael Martens2, Chaitra Badve1,3, and Mark Griswold1
1Radiology, Case Western Reserve University, Cleveland, OH, United States, 2Physics, Case Western Reserve University, Cleveland, OH, United States, 3Radiology, University Hospitals Cleveland Medical Center, Cleveland, OH, United States
In this study, we introduced a new deep learning method to take advantage of both radiologists’ expertise and multi-parametric MR Fingerprinting data to improve annotation accuracy in MRI dataset. A U-Net based convolutional neural network was adopted and each dataset was evaluated multiple times using different combinations of training dataset. Our initial results obtained from a brain tumor dataset demonstrates that the developed method could effectively identify mislabeled tissues and improve annotation accuracy.
|
||
1346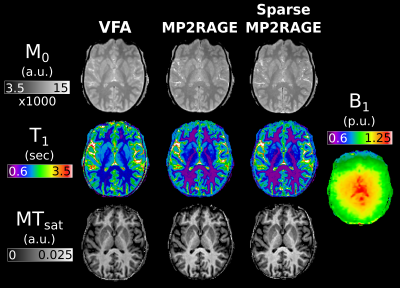 |
52 | Efficient MTsat mapping using sparse MP2RAGE for T1 and M0 measurement with B1+ inhomogeneity correction
Christopher D Rowley1,2, Ilana R. Leppert1, Jennifer S.W. Campbell1, Mark C. Nelson1, G Bruce Pike3, and Christine L Tardif1,2,4
1McConnell Brain Imaging Centre, McGill University, Montreal, QC, Canada, 2Neurology and Neurosurgery, McGill University, Montreal, QC, Canada, 3Hotchkiss Brain Institute and Departments of Radiology and Clinical Neuroscience, University of Calgary, Calgary, AB, Canada, 4Biomedical Engineering, McGill University, Montreal, QC, Canada In this study we investigate the use of MP2RAGE derived M0 and T1 maps for use in MTsat imaging. These values are compared against maps calculated from the traditional VFA approach. Additionally, the MP2RAGE approach is evaluated with and without sparse sampling and compressed sensing reconstruction. We show that VFA produces elevated T1 values, and consequently lower MTsat compared to MP2RAGE approaches. A model-based ΔB1+ correction was able to remove the dependence of MTsat on ΔB1+. The sparse MP2RAGE provided high quality maps, leading to a full MTsat protocol that was 32% faster than the traditional approach. |
||
1347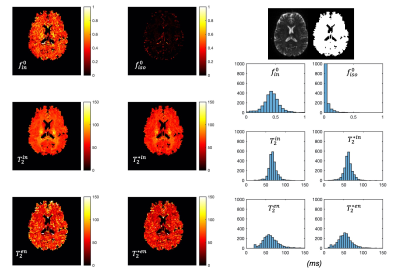 |
53 | Simultaneous mapping of compartment-specific T2 and T2* with Diffusion-PEPTIDE imaging
Ting Gong1, Merlin J. Fair2, Kawin Setsompop2,3, and Hui Zhang1
1Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 2Radiological Sciences Laboratory, Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States
Inspired by recent developments in combined relaxometry-diffusion imaging and growing interests in T2* imaging, this study achieves simultaneous compartmental T2 and T2* mapping for the first time. We take advantage of the recently developed diffusion-PEPTIDE imaging technique, which allows for efficient acquisition of diffusion datasets with varying T2/T2* contrast. A relaxometry-diffusion combined microstructure model and a robust multi-stage fitting approach are developed to enable compartmental T2 and T2* mapping. This technique could be a new tool for future neuroimaging studies benefiting from quantification of T2-T2*-diffusion, such as studying iron content related development and pathology.
|
||
1348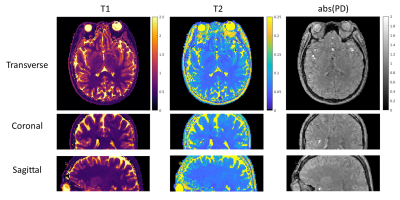 |
54 | 3D MR-STAT: towards a fast multi-parametric protocol with increased SNR
Hongyan Liu1, Oscar van den Heide1, Miha Fuderer1, Cornelis A.T. van den Berg1, and Alessandro Sbrizzi1
1Computational Imaging Group for MR diagnostics & therapy, Center for Image Sciences, UMC Utrecht, Utrecht, Netherlands
MR-STAT is a framework for simultaneous mapping of quantitative MR parameters from single short scans. In this work, we extend the current 2D Cartesian MR-STAT sequence to 3D acquisitions, in order to achieve higher SNR and isotropic resolution with a scan lasting few minutes. In particular, we implement a prototype 3D MR-STAT sequence for whole brain coverage with isotropic resolution, and test it on gel phantom and in-vivo brain data. Acceleration strategy of the 3D sequence is proposed, and the corresponding retrospective undersampling of the measured in-vivo data is reconstructed.
|
||
1349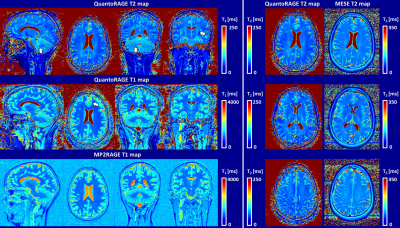 |
55 | Simultaneous T1 and T2 mapping with QuantoRAGE: a new MP2RAGE variant using T2-prepared inversion Video Permission Withheld
Gabriele Bonanno1,2,3, José P. Marques4, Tobias Kober5,6,7, and Tom Hilbert5,6,7
1Siemens Healthcare AG, Bern, Switzerland, 2Magnetic Resonance Methodology, Institute of Diagnostic and Interventional Neuroradiology, University of Bern, Bern, Switzerland, 3Translational Imaging Center, sitem-insel, Bern, Switzerland, 4Donders Institute for Brain Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 5Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 6Department of Radiology, University Hospital (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland, 7LTS5, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland T1 and T2 relaxometry provide important quantitative information and can serve as imaging biomarkers thanks to their sensitivity to pathology. However, quantitative imaging requires long scan times for high-resolution whole-brain coverage. Magnetization-prepared approaches combined with fast sequences allow for high isotropic resolution, but they are often biased due to other relaxation mechanisms than the one being probed. To account for this effect, we introduce QuantoRAGE: a new method that uses a T2-prepared inversion within an accelerated MP2RAGE sequence for simultaneous T1 and T2 mapping. Preliminary tests demonstrate the feasibility to obtain high-resolution simultaneous T1 and T2 relaxometry at 3T. |
||
1350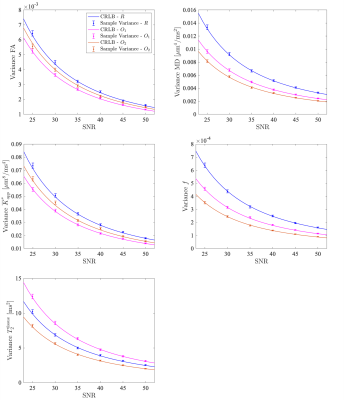 |
56 | Optimal acquisition settings for simultaneous diffusion kurtosis, free water fraction and T2 estimation
Vincenzo Anania1,2, Ben Jeurissen1,3, Jan Morez1, Annemieke E. Buikema1, Thibo Billiet2, Jan Sijbers1,3, and Arnold J. den Dekker1,3
1imec-Vision Lab, Dept. of Physics, University of Antwerp, Antwerp, Belgium, 2icometrix, Leuven, Belgium, 3µNEURO Research Centre of Excellence, University of Antwerp, Antwerp, Belgium
Fitting the diffusion kurtosis imaging free water elimination model (DKI-FWE) to diffusion MRI data represents an ill-conditioned problem. Fortunately, the conditioning of the model fitting can be improved by explicitly modeling the T2 relaxation dependency of the signal. As a benefit, diffusion and kurtosis metrics robust to partial volume effects can be estimated with conventional techniques. In this work, we use Cramér-Rao lower bound (CRLB) theory to identify optimal acquisition settings that maximize the precision of the model parameter estimates.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.