Online Gather.town Pitches
Processing & Analysis II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
 |
4385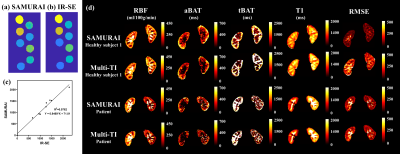 |
1 | Saturated Multi-delay Arterial Spin Labeling (SAMURAI) Technique for Simultaneous Perfusion and T1 Quantification of Kidneys
Zihan Ning1, Shuo Chen1, Zhensen Chen2, Hualu Han1, Huiyu Qiao1, Rui Shen1, Peng Wu3, and Xihai Zhao1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine Tsinghua University, Beijing, China, 2Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 3Philips Healthcare, Shanghai, China
We proposed a Saturated Multi-delay Arterial Spin Labeling (SAMURAI) technique with a correspondingly modified kinetic model to achieve simultaneous acquisitions of RBF, aBAT, tBAT and T1 map in kidney with a single scan. SAMURAI provided T1 map with excellent correlation (R2=0.976) compared with IR-SE. Compared with multi-TI FAIR, SAMURAI provided equally reliable ASL and T1 quantification results (ICC: 0.875-0.958) with excellent scan-rescan repeatability (ICC: 0.905-0.992) and significantly reduced scan time (45’ vs 4’6’’ for 9 TIs).
|
|
 |
4386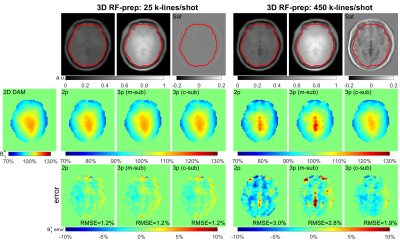 |
2 | Ultrafast B1 Mapping with RF-Prepared 3D FLASH Acquisition: Correcting the Bias due to T1-Induced k-Space Filtering Effect
Dan Zhu1,2 and Qin Qin1,2
1F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 2The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States
The traditional RF-prepared B1 mapping technique consists of one scan with an RF preparation module for FA-encoding and a second scan without this module for normalizing. To reduce the T1-induced k-space filtering effect, this method is limited to 2D FLASH acquisition with a two-parameter method. Based on the point spread function analysis of FLASH readout, a novel 3D RF-prepared three-parameter method for B1-mapping is proposed to correct the T1-induced quantification bias. The proposed technique was compared with existing methods in the brain, breast, and abdomen and demonstrated with high accuracy for ultrafast B1 mapping.
|
|
4387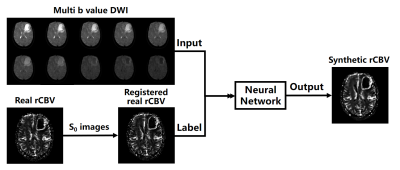 |
3 | Generation of DSC-MRI derived relative CBV maps from IVIM-MRI data
Lu Wang1, Zhen Xing2, Qinqin Yang1, Congbo Cai1, Zhong Chen1, Dairong Cao2, Zhigang Wu3, and Shuhui Cai1
1Department of Electronic Science, Xiamen University, Xiamen, Fujian, China, 2Department of Radiology, First Affiliated Hospital of Fujian Medical University, Fuzhou, Fujian, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China
Dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) derived relative cerebral blood volume (rCBV) is a valuable diagnosis biomarker. However, the injection of gadolinium-based contrast agent (GBCA) in DSC-MRI acquisition is prone to cause adverse effects. In this study, a rCBV generation method was proposed based on deep neural network and intravoxel incoherent motion magnetic resonance imaging (IVIM-MRI) data. Consistency analysis shows that the rCBV maps generated from our proposed method are of high consistency with the realistic ones, implying that the proposed method has the potential to obtain DSC-MRI derived rCBV maps without GBCA injection.
|
||
4388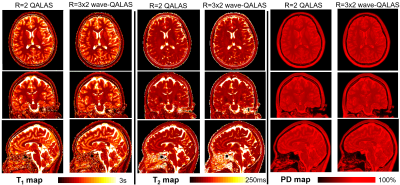 |
4 | Time-efficient, high resolution 3T whole brain relaxometry using 3D-QALAS with wave-CAIPI readouts
Borjan Gagoski1,2, Jaejin Cho2,3, Zijing Zhang4, Tae Hyung Kim2,3, Wei-Ching Lo5, Daniel Polak6, Marcel Warntjes7, Stephen Cauley2,3, Kawin Setsompop8,9, P. Ellen Grant1,2, and Berkin Bilgic2,3
1Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 4State Key Laboratory of Modern Optical Instrumentation, College of Optical Science and Engineering, Zhejiang University, Hangzhou, Zhejiang, China, 5Siemens Medical Solutions USA, Inc., Charlestown, MA, United States, 6Siemens Healthcare GmbH, Erlangen, Germany, 7SyntheticMR AB, Linköping, Sweden, 8Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 9Department of Radiology, Stanford University, Stanford, CA, United States Volumetric, high resolution, quantitative mapping of brain tissues’ relaxation properties is hindered by long acquisition times and signal-to-noise (SNR) challenges. This study, for the first time, combines the time-efficient wave-CAIPI readouts with the 3D-Quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) scheme, enabling full brain quantitative T1, T2 and proton density (PD) maps at 1.15 mm isotropic voxels in only 3 minutes at R=3x2 acceleration. When tested in both the ISMRM/NIST phantom and 7 healthy volunteers, the quantitative maps using the accelerated protocol showed excellent agreement against those obtained from conventional 3D-QALAS at RGRAPPA=2. |
||
 |
4389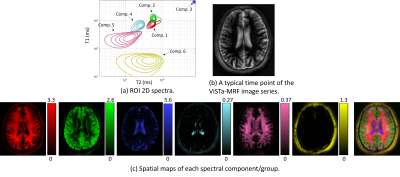 |
5 | Estimating multicomponent 2D relaxation spectra with a ViSTa-MR fingerprinting acquisition
Yunsong Liu1, Congyu Liao2, Daeun Kim3, Kawin Setsompop2, and Justin P. Haldar3
1Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Stanford University, Stanford, CA, United States, 3University of Southern California, Los Angeles, CA, United States
Multidimensional relaxation correlation spectroscopic imaging methods have demonstrated powerful capabilities to resolve subvoxel microstructure. In this work, we perform T1-T2 relaxation correlation spectroscopic imaging using a sequence that combines MR fingerprinting with a ViSTa preparation module to enhance sensitivity to short-T1 components. We demonstrate theoretically and empirically that this approach has advantages over MR fingerprinting without ViSTa. Empirical results demonstrate the ability to identify at least 6 anatomically plausible tissue components, including a short-T1 component that was not previously resolved when using MR fingerprinting without ViSTa. A novel generalized ADMM algorithm is also proposed that substantially improves computational efficiency.
|
|
 |
4390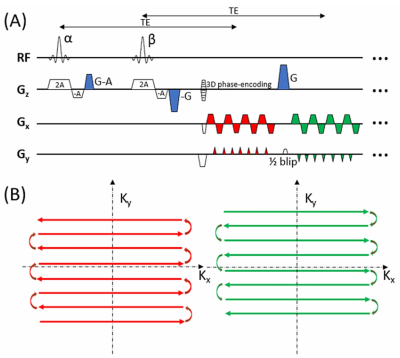 |
6 | Three-dimensional echo-shifted echo planar imaging with simultaneous blip-up and blip-down acquisitions for correcting geometric distortion
Kaibao Sun1, Zhifeng Chen2,3, Guangyu Dan1,4, Qingfei Luo1, Alessandro Scotti1, Lirong Yan5,6, and Xiaohong Joe Zhou1,4,7
1Center for MR Research, University of Illinois at Chicago, Chicago, IL, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Charlestown, MA, United States, 4Department of Biomedical Engineering, University of Illinois at Chicago, Chicago, IL, United States, 5USC Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States, 6Department of Neurology, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States, 7Departments of Radiology and Neurosurgery, University of Illinois at Chicago, Chicago, IL, United States
Geometric distortion is a prevalent image artifact in echo planar imaging (EPI). In a method known as BUDA (blip-up/down acquisition), k-space model-based reconstruction of two EPI datasets acquired separately with opposing phase-encoding polarities has been shown to reduce image distortions. A main disadvantage is that the two-shot acquisition strategy doubles the scan time and increases vulnerability to motion. We herein introduce a novel sequence – 3D echo-shifted EPI with BUDA (esEPI-BUDA) – to address the aforementioned issues by integrating the two acquisitions into one shot. We have successfully applied the 3D esEPI-BUDA technique to functional MRI.
|
|
 |
4391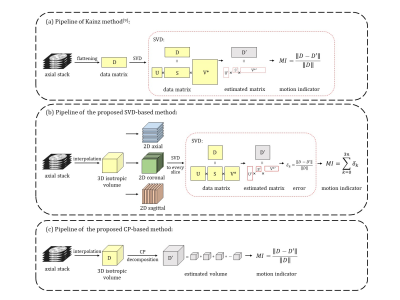 |
7 | A Motion Assessment Method for Fetal Brain MRI Based on 2D-SVD and 3D-CP Decomposition
Haoan Xu1, Wen Shi1,2, Jiwei Sun1, Cong Sun3, Guangbin Wang3,4, Yi Zhang1, and Dan Wu1
1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 2Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Radiology, Cheeloo College of Medicine, Shandong Provincial Hospital, Shandong University, Jinan, China, 4Department of Radiology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China
Slice-to-volume registration and super-resolution reconstruction is commonly used to generate 3D volumes of the fetal brain from 2D stacks in multiple orientations. The current pipeline requires selecting the stack with minimal motion as a reference for registration. We proposed a motion assessment method that automatically determines the reference stack based on CANDECOMP/PARAFAC decomposition. This method is sensitive to motion across slices compared to other state-of-the-art methods. Combining motion assessment with the existing fetal brain MRI processing pipeline improved the reconstruction quality and the success rate.
|
|
4392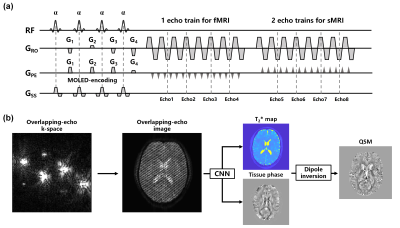 |
8 | Single-shot simultaneous T2* mapping and QSM using overlapping-echo acquisition for quantitative structural and functional MRI
Qinqin Yang1, Lingceng Ma1, Shuhui Cai1, Hongjian He2, Zhong Chen1, Jianhui Zhong2,3, and Congbo Cai1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Center for Brain Imaging Science and Technology, College of Biomedical Engineering and Instrumental Science, Zhejiang University, Zhejiang, China, 3Department of Imaging Sciences, University of Rochester, Rochester, NY, United States
A quantitative magnetic resonance imaging (MRI) sequence GRE-MOLED based on overlapping-echo detachment is implemented to simultaneously achieve distortion-free T2* mapping and quantitative susceptibility mapping (QSM) in a single shot. The accuracy of quantitative structural MRI results is verified by comparison with the results from multi-shot method in in-vivo experiments. We also evaluate the technique through a statistical analysis of fMRI experiments with vision and motion tasks and demonstrate that it achieves higher BOLD sensitivity than conventional method.
|
||
4393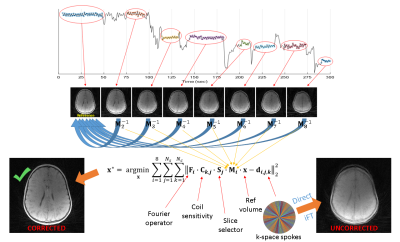 |
9 | Motion-corrected radial head MRI without any navigator, data redundancy or external tracking
Cihat Eldeniz1, Paul K. Commean1, Parna E. Boroojeni1, Jamal Derakhshan1, Yan Yan2, Udayabhanu Jammalamadaka3, Manu S. Goyal1, Gary Skolnick4, Kamlesh B. Patel4, and Hongyu An5
1Mallinckrodt Institute of Radiology, Washington University in St. Louis, SAINT LOUIS, MO, United States, 2Public Health Sciences, Washington University in St. Louis, SAINT LOUIS, MO, United States, 3Office of Research, Rice University, Houston, TX, United States, 4Division of Plastic and Reconstructive Surgery, Washington University in St. Louis, SAINT LOUIS, MO, United States, 5Mallinckrodt Institute of Radiology, Washington University in St. Louis, St. Louis, MO, United States
Patient motion is a common problem in MRI. Both prospective and retrospective schemes can be inefficient if they use navigators. Therefore, self-navigation without any data redundancy is preferable. In this study, we propose a simple and robust motion detection scheme that is free from stringent assumptions. This is especially important while imaging the head because the motion patterns are unpredictable. The corrected and uncorrected images were reviewed by two neuroradiologists in terms of osseous distinction (in reference to CT), sharpness and artifact-freeness. The method is able to increase osseous distinction and sharpness without increasing artifacts. This offers substantial clinical utility.
|
||
4394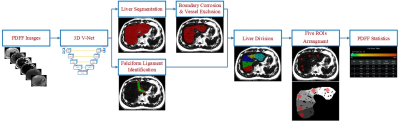 |
10 | Deep Learning based automatic ROI sampling approach for the measurement of liver PDFF
Xinxin Xu1, Yihuan Wang2, Shuheng Zhang3, Xiang Chen3, Yang Li3, Ke Wu3, Jianmin Yuan4, and Fuhua Yan2
1Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 2Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 3United Imaging Healthcare, Shanghai, China, 4Central Research Institute, United Imaging Healthcare, Shanghai, China Hepatic proton density fat fraction (PDFF) measurement plays an important role in the assessment of chronic diffuse liver diseases, while it is always time consuming and lack of good reproducibility and repeatability. To address this problem, we introduced a five-ROI sampling approach based on a convolutional neural network that provided high dice coefficient (DC) of whole liver segmentation, good correlation and quicker compared with manual operation. |
||
4395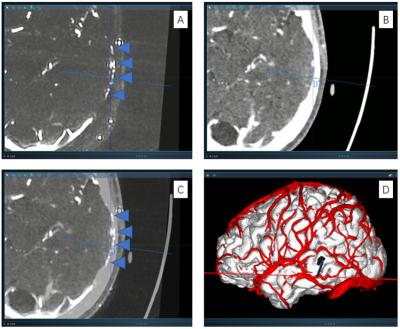 |
11 | Utilization of high resolution and low velocity encoding PCA with highly accelerated compressed sensing for preoperative SEEG planning Video Permission Withheld
Qiangqiang Liu1,2, Shuheng Zhang3, Jiwen Xu1,2, Jiachen Zhu3, Yiwen Shen4, Wenzhen Chen1,2, Xiaolai Ye1,2, Dong Wang3, and Jianmin Yuan5
1Department of Neurosurgery, Clinical Neuroscience Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 2Clinical Neuroscience Center, Ruijin Hospital Luwan Branch, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 3United Imaging Healthcare, Shanghai, China, 4Department of Radiology, Ruijin Hospital Luwan Branch, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 5Central Research Institute, United Imaging Healthcare, Shanghai, China
It is essential to avoid small vessels during stereo-electroencephalography (SEEG) electrode implantation. In this study, we proposed a 6-fold Compressed sensing accelerated, 5cm/s Low velocity encoded, 0.75mm Isotropic resolution Phase contrast Magnetic Resonance Angiography (CLIP-MRA). In CLIP-MRA, the compressed sensing based acceleration method was shown to achieve better image quality or shorter scan duration compared to parallel imaging based acceleration. CLIP-MRA was able to display not only cortical arteries and veins simultaneously, but also vessels in the skull. Safety and effectiveness of CLIP-MRA utilized preoperative SEEG planning were evaluated on a small patient cohort.
|
||
4396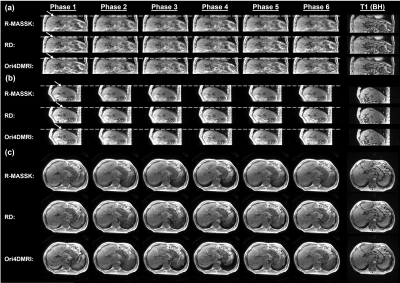 |
12 | Retrospective Motion Artifacts Suppression by Simulated radial K-space (R-MASSK) for 3D view-sharing sequence acquired abdominal 4D-MRI
Yat Lam Wong1, Hing Chiu Chang2, Tian Li1, and Jing Cai1
1Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 2Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong, Hong Kong
3D view-sharing sequence has been demonstrated its great promise for abdominal 4D-MRI due to its high temporal resolution and availability in clinical scanners. However, the sub-optimal use of these sequence in free-breathing situation deteriorates the image quality by ghost artifacts induced by respiration. We propose a novel technique, Retrospective Motion Artifacts Suppression by Simulated radial K-space (R-MASSK), to suppress motion artifacts and to increase image quality of the 4D-MRI acquired by the 3D view-sharing sequences, TRICKS and TWIST, through simulated radial k-space resampling.
|
||
4397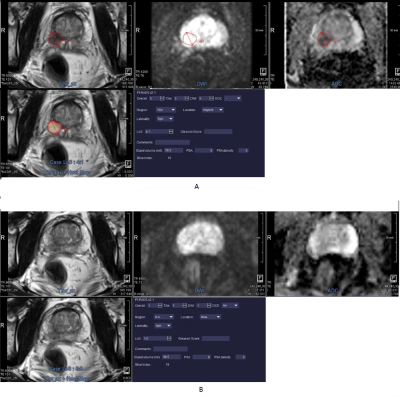 |
13 | MRI Based Automated Deep-Learning System in the Assessment of Prostate Cancer: Comparison of Advanced Zoomed DWI and Conventional Technique
Lei Hu1, Caixia Fu2, Robert Grimm3, Heinrich von Busch4, Thomas Benkert 3, and Jun-gong Zhao1
1Department of Diagnostic and Interventional Radiology, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai, China, 2MR Application Development, Siemens Shenzhen Magnetic Resonance Ltd, Shenzhen, China, 3MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 4Innovation Owner Artificial Intelligence for Oncology, Siemens Healthcare GmbH, Erlangen, Germany
Prostate cancer (PCa) detection by an automated deep-learning computer-aided diagnosis (DL-CAD) system using advanced zoomed diffusion-weighted imaging (z-DWI) and full-field-of-view DWI (f-DWI) were compared. The DL-CAD system using z-DWI performed significantly better for PCa detection accuracy per patient (AUC: 0.937 vs. 0.871; P=0.002) and had significantly higher PCa lesion detection accuracy per lesion compared to f-DWI (AUC: 0.912 vs. 0.833; P=0.003). Given this, use of z-DWI can improve the performance of the DL-CAD system for PCa detection.
|
||
4398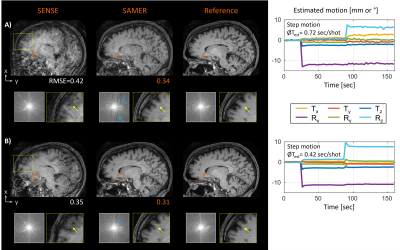 |
14 | Linear+: An optimized sequence reordering for robust scout accelerated retrospective motion estimation and correction
Daniel Polak1,2, Daniel Nicolas Splitthoff1, Bryan Clifford3, Lo Wei-Ching3, Azadeh Tabari4, Susie Huang4, John Conklin4, Lawrence L. Wald2, and Stephen Cauley2
1Siemens Healthcare GmbH, Erlangen, Germany, 2A. A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Siemens Medical Solutions, Malvern, PA, United States, 4Department of Radiology, Massachusetts General Hospital, Boston, MA, United States
Distributed sequence reorderings for 3D multi-shot acquisitions ensure overlap with the central k-space in every shot. This improves the stability of navigator-free retrospective motion estimation but often reduces the robustness of the image reconstruction. In this work, we have developed and optimized a novel sampling strategy (Linear+) that extends the standard sequential sequence ordering with a small number of additional calibration samples acquired near the k-space center. In simulation and in vivo, Linear+ enabled accurate motion parameter estimation and correction across multiple motion patterns while providing improved image homogeneity and spatial resolution compared to a distributed reordering.
|
||
4399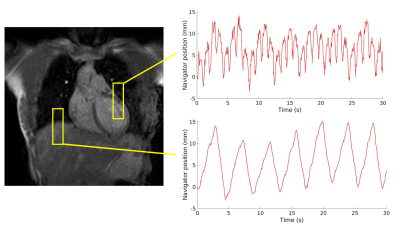 |
15 | Joint cardiorespiratory 1D navigator triggering using left ventricular motion: improving T2w-FSE imaging for thoracic radiotherapy
Aart van Bochove1, Katrinus Keijnemans1, Osman Akdag1, Pim Borman1, Martin Fast1, and Astrid van Lier1
1Department of Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands
For MR-guided radiotherapy treatment planning of the thorax, cardiorespiratory triggered T2-weighted MRI images are desired. We investigate the usage of a navigator placed on the left ventricle to do cardiorespiratory triggering, and compare it to images obtained by placing the navigator on the liver-lung interface to do respiratory triggering. We scanned 5 healthy volunteers, making high-resolution scans with both navigators, and cine and 4D-MRI scans for reference. We found that cardiorespiratory triggering based on left ventricular motion is possible, and increases image quality, without increasing scan time and losing geometrical respiratory information.
|
||
4400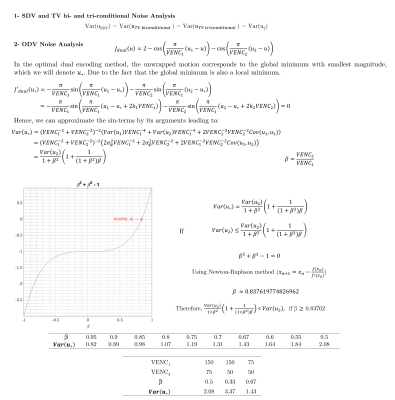 |
16 | A Review and Comparison of Unwrapping Methods in Dual Velocity-Encoding MRI
Pamela Franco1,2,3, Liliana Ma4,5, Susanne Schnell6, Michael Markl4,5, Cristobal Bertoglio7, and Sergio Uribe1,2,8
1Biomedical Imaging Center, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 2Millennium Nucleus for Cardiovascular Magnetic Resonance, Santiago, Chile, 3Electrical Engineering Department, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 4Department of Biomedical Engineering, McCormick School of Engineering, Northwestern University, Chicago, IL, United States, 5Department of Radiology, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States, 6Institut für Physik, Universität Greifswald, Greifswald, Germany, 7Bernoulli Institute, University of Groningen, Groningen, Netherlands, 8Radiology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile
4D flow MRI data may be inaccurate due to phase wrapping when using lower VENC than the maximum actual velocity. Accuracy can also be affected by reduced velocity-to-noise ratio (VNR) when using a too high VENC. Dual-VENC approaches have been proposed to unwrap velocity-aliasing artifacts and increase VNR, providing more reliable measurements and allowing more flexibility in the selection of VENC. Dual-VENC methods enable acquisitions of 4D flow MRI data with high dynamic range and without velocity aliasing. The purpose of this study is to compare the performance of some of these methods.
|
||
4401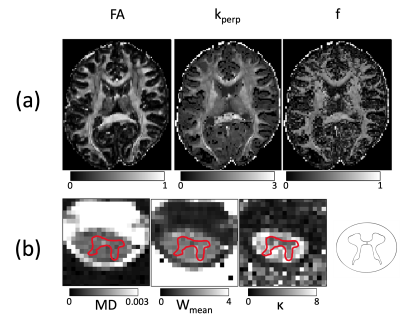 |
17 | ACID - an open-source, bids compatible software for brain and spinal cord dMRI: preprocessing, DTI/DKI, biophysical modelling
Björn Fricke1, Gergely David1,2, Jan Malte Oeschger1, Patrick Freund2, Lars Ruthotto3, Karsten Tabelow4, and Siawoosh Mohammadi1,5
1Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 2Balgrist University Hospital, University of Zurich, Zürich, Switzerland, 3Department of Mathematics, Emory University, Atlanta, GA, United States, 4Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 5Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
The ACID-Toolbox is an open-source toolbox for brain and spinal-cord diffusion data. It enables the preprocessing and creation of DTI/DKI, and biophysical parameter-maps in one single pipeline. Preprocessing covers correction for eddy-currents and motion-artefacts, susceptibility distortions and adaptive denoising. For model-fitting, the toolbox offers several algorithms to estimate the diffusion or the kurtosis tensors as well as estimation of biophysical parameters. ACID is integrated into the batch system of the Statistical-Parametric-Mapping (SPM12) software for the analysis of neuroimaging data and thus benefits from associated spatial and statistical processing modules. Additionally, ACID is BIDS compatible but also applicable to non-BIDS-conform data.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.