Online Gather.town Pitches
Image Reconstruction I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
 |
3473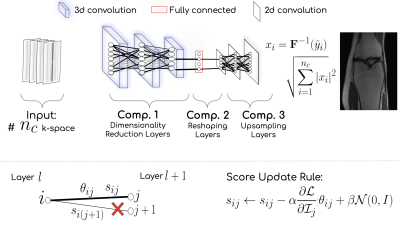 |
1 | A domain-agnostic MR reconstruction framework using a randomly weighted neural network
Arghya Pal1 and Yogesh Rathi1
1Department of Psychiatry, Harvard Medical School, Boston, MA, United States
Can a random weighted deep network structure encode informative cues to solve the MR reconstruction problem from highly under-sampled k-space measurements? Trained networks update the weights at training time, while untrained networks optimize the weights at inference time. In contrast, our proposed methodology selects an optimal subnetwork from a randomly weighted dense network to perform MR reconstruction without updating the weights - neither at training time nor at inference time. The methodology does not require ground truth data and shows excellent performance across domains in T1-weighted (head, knee) images from highly under-sampled multi-coil k-space measurements.
|
|
3474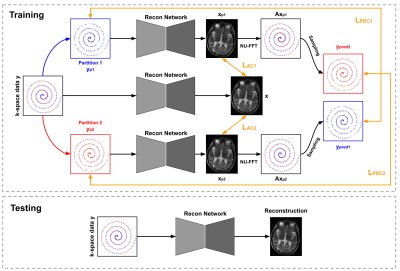 |
2 | Dual-domain Self-supervised Learning for Accelerated MRI Reconstruction
Bo Zhou1,2, Jo Schlemper2, Seyed Sadegh Mohseni Salehi 2, Neel Dey2,3, Kevin Sheth1, Chi Liu1, James Duncan1, and Michal Sofka2
1Yale University, New Haven, CT, United States, 2Hyperfine Research, Guilford, CT, United States, 3New York University, New York, NY, United States
We present a self-supervised approach for accelerated non-uniform MRI reconstruction, which leverages self-supervision in k-space and image domains. We evaluated the performance on both simulation and real data, where fully sampled data is unavailable. The experimental results on a non-uniform MRI dataset demonstrate that the proposed method can generate reconstruction that approaches the accuracy of the fully supervised reconstruction. Furthermore, we show that the approach can be applied to highly challenging real-world clinical MRI reconstruction acquired on a low-field (64 mT) MRI scanner with no data available for supervised training while demonstrating improved perceptual quality as compared to traditional reconstruction.
|
||
3475 |
3 | Dynamic spectral modes of resting-state BOLD time courses in white matter
Muwei Li1, Yurui Gao1, Adam W Anderson1, Zhaohua Ding1, and John C Gore1
1Vanderbilt University Institute of Imaging Science, Nashville, TN, United States
We investigated voxel-wise spectrograms derived from the time-varying spectral patterns of BOLD signals in white matter. Finite-window spectra are non-stationary but may be categorized into five distinct modes that recur over time. Close scrutiny of the signal profiles reveals distinct spatial distributions of the occurrences and durations of these modes, and such distributions are highly consistent across individuals. In addition, two communities of white matter voxels may be identified according to a hierarchical arrangement of transitions among modes. Our findings reveal the non-stationary nature of BOLD spectral patterns, and provide a novel spatial-temporal-frequency characterization of resting-state signals in white matter.
|
||
3476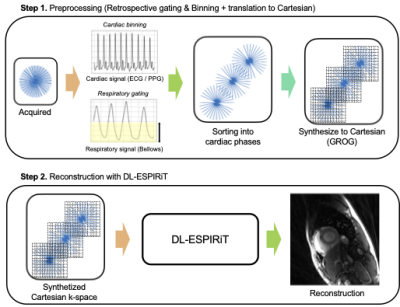 |
4 | Accelerated free-breathing radial cine imaging via GROG-interpolated DL-ESPIRiT
Kanghyun Ryu1, Zhitao Li1, Christopher M. Sandino2, and Shreyas S. Vasanawala1
1Radiology, Stanford University, Stanford, CA, United States, 2Electrical Engineering, Stanford University, Stanford, CA, United States Deep Learning based reconstruction methods have been vastly explored for accelerating Cartesian-based cardiac cine imaging via using unrolled neural networks. However, for non-Cartesian trajectories such as radial, these networks require substantial modifications (i.e., NUFFT-based data consistency) and requires collecting separate radial-based dataset, which may not be common in the clinics. Here, we investigate a method to transfer the radial k-space data to the Cartesian domain using GROG-based interpolation. We show that DL-ESPIRiT trained with Cartesian cine dataset (with pseudo radial-like under sampling pattern) can be generalizable to reconstruct actual accelerated radial cine acquired on a scanner. |
||
3477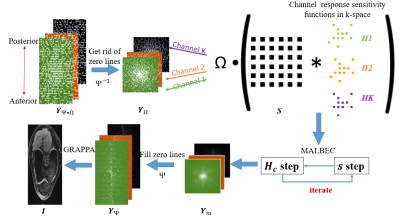 |
5 | Combination of Multichannel Blind Deconvolution (MALBEC) and GRAPPA for Highly Accelerated 3D Imaging
Ruiying Liu1, Jee Hun Kim2, Chaoyi Zhang1, Hongyu Li1, Peizhou Huang1, Xiaojuan Li2, and Leslie Ying1
1Department of Biomedical Engineering, Department of Electrical Engineering, University at Buffalo, Buffalo, NY, United States, 2Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States
Most parallel imaging methods require calibration data for reconstruction. Low-rank-based methods allow calibration-free reconstruction from randomly undersampled, multi-channel data. This abstract presents a novel reconstruction method to combine multichannel blind deconvolution (MALBEC), a calibration-free method, and GRAPPA, a calibration-based method for highly accelerated imaging. The method sequentially performs MALBEC and GRAPPA with specially designed sampling masks such that the benefits of low-rank structure and the availability of calibration data can be utilized jointly. Our results demonstrate that the proposed method can achieve an acceleration factor that is the product of the factors achieved by MALBEC and GRAPPA alone.
|
||
3478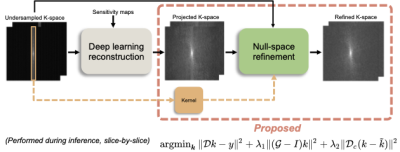 |
6 | K-space refinement method for DL-based MR reconstruction by regularizing k-space null space constraint with auto-calibrated kernel
Kanghyun Ryu1, Cagan Alkan2, and Shreyas S. Vasanawala1
1Radiology, Stanford University, Stanford, CA, United States, 2Electrical Engineering, Stanford, Stanford, CA, United States
In this study, we propose a novel refinement method using auto-calibrated k-space null-space kernel to reduce the k-space errors and enable reconstruction of improved high-frequency image details and textures. The refinement algorithm can be easily plugged in after DL-based reconstructions. We show that our method enables the reconstruction of sharper images with significantly improved high-frequency components measured by HFEN and GMSD while maintaining overall error in the image measured by PSNR and SSIM.
|
||
3479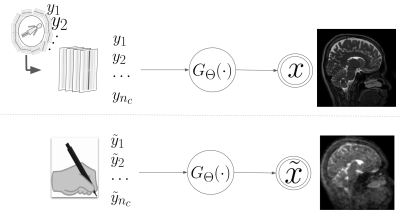 |
7 | Understanding and Reducing structural bias in deep learning-based MR reconstruction
Arghya Pal1 and Yogesh Rathi2
1Department of Psychiatry, Harvard Medical School, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
Deep learning methods are increasingly being used for accelerating magnetic resonance imaging (MRI) acquisition and reconstruction strategies. However, it is important to understand whether or not deep learning models have an inherent structural bias that may effectively create concerns in real-world settings. In this abstract, we show a strategy to understand and then reduce structural prior bias in deep models. The proposed approach decouples the spurious structural bias (prior) of a deep learning model by intervening in the input. Our proposed debiasing strategy is fairly robust and can work with any pre-trained deep learning MR reconstruction model.
|
||
3480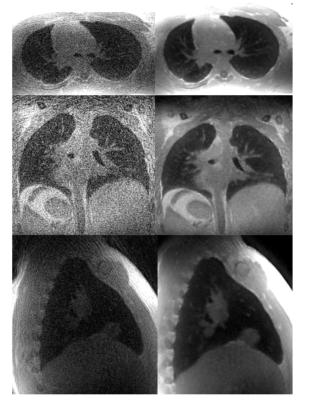 |
8 | Memory Efficient Model Based Reconstruction for Volumetric Non-Cartesian Acquisitions using Gradient Checkpointing and Blockwise Learning
Zachary Miller1 and Kevin Johnson1
1University of Wisconsin-Madison, Madison, WI, United States
The goal of this work is to reconstruct high resolution, volumetric non-cartesian acquisitions using model based deep learning. This is a challenging problem because unlike cartesian acquisitions and hybrid trajectories like stack of spirals that can be decoupled into 2D sub problems, fully non-cartesian trajectories require the complete 3D volume for data-consistency pushing GPU memory capacity to its limits even using cluster computing. Here, we investigate blockwise learning, a method that has the memory saving benefits of patch-based methods while maintaining data-consistency in k-space. We demonstrate the feasibility of this method by reconstructing high resolution pulmonary non-contrast and MRA acquisitions.
|
||
3481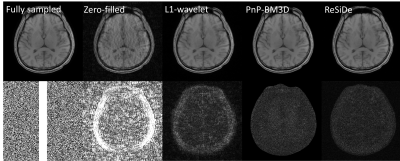 |
9 | Image Reconstruction with a Self-calibrated Denoiser
Sizhuo Liu1, Philip Schniter1, and Rizwan Ahmad1
1The Ohio State University, Columbus, OH, United States
Plug-and-play (PnP) methods can reconstruct images by employing iterative algorithms that leverage the knowledge of the forward model and a sophisticated denoiser. The performance of PnP can be improved by utilizing an application-specific denoiser. However, training such denoisers may not be feasible for many MRI applications. Here, we describe a PnP-inspired method that does not require data beyond the single, incomplete set of measurements. The proposed method, called recovery with a self-calibrated denoiser (ReSiDe), trains the denoiser from the patches of the image being recovered. For validation, ReSiDe is applied to T1-weighted brain and myocardial first-pass perfusion data.
|
||
3482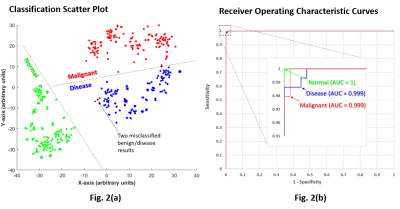 |
10 | A single artificial neural network solution to detect pancreatic and lung cancer from high-resolution 1H MR plasma/serum spectra
Meiyappan Solaiyappan1, Santosh Kumar Bharti1, Mohamad Dbouk2, Wasay Nizam3, Malcolm V. Brock3,4, Michael G. Goggins2,4,5, and Zaver M. Bhujwalla1,2,6
1Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Department of Pathology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Surgery, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 4Department of Oncology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5Department of Medicine, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 6Radiation Oncology and Molecular Radiation Sciences, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Early detection of cancers using blood-based analytes for routine screening is a rapidly advancing area. Here, we developed a neural-network based solution to detect pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung cancer (NSCLC) with high sensitivity and specificity using human plasma and serum samples to discriminate between subjects with no known pancreatic or lung disease, subjects with benign disease and subjects with PDAC or NSCLC.
|
||
3483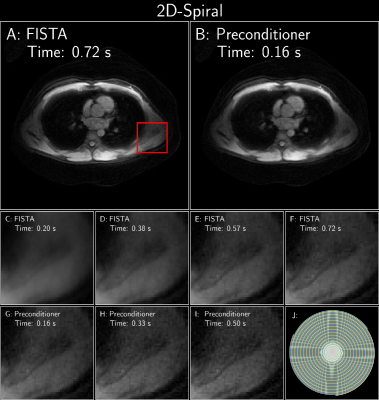 |
11 | Polynomial Preconditioning for Accelerated Convergence of Proximal Algorithms including FISTA
Siddharth Srinivasan Iyer1,2, Frank Ong3, and Kawin Setsompop2,3
1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States
This work aims to accelerate the convergence of proximal gradient algorithms (like FISTA) by designing a preconditioner using polynomials that targets the eigenvalue spectrum of the forward linear model to enable faster convergence. The preconditioner does not assume any explicit structure and can be applied to various imaging applications. The efficacy of the preconditioner is validated on four varied imaging applications, where it seen to achieve 2-4x faster convergence.
|
||
3484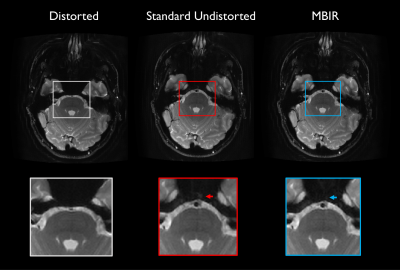 |
12 | Model-based PSF-encoded multi-shot EPI reconstruction with low-rank constraints
Nolan K Meyer1,2, Myung-Ho In2, Daehun Kang2, Yunhong Shu2, John Huston III2, Matt A Bernstein2, and Joshua D Trzasko2
1Mayo Clinic Graduate School of Biomedical Sciences, Rochester, MN, United States, 2Department of Radiology, Mayo Clinic, Rochester, MN, United States
Echo planar imaging (EPI) is commonly used clinically for its speed, but is sensitive to non-idealities including system field inhomogeneity and subject-specific susceptibility effects. Multi-shot techniques encoding an auxiliary point-spread-function (PSF)-encoding dimension provide robustness to off-resonance effects, with demonstrated potential for diffusion and anatomic applications, but incur scan time penalties motivating acquisition and reconstruction strategies to increase acceleration. This work proposes a comprehensive model-based iterative reconstruction framework for anatomic PSF-encoded EPI scans incorporating low-rank constraints, directly reconstructing undistorted images from undersampled data. Advantages are demonstrated using brain MRI data acquired on a compact 3T MRI system.
|
||
3485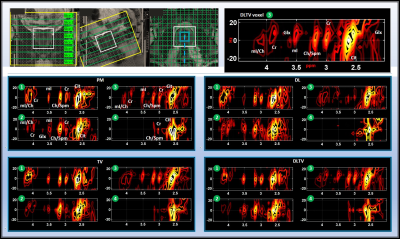 |
13 | Accelerated Echo Planar J-Resolved Spectroscopic Imaging in Prostate Cancer and A Hybrid Dictionary Learning-Total Variation Reconstruction
Ajin Joy1, Rajakumar Nagarajan1, Andres Saucedo1, Zohaib Iqbal1, Manoj K Sarma1, Neil E Wilson1, Ely R Felker1, Robert E Reiter2, Steven S Raman1, and M Albert Thomas1
1Radiological Sciences, University of California-Los Angeles, Los Angeles, CA, United States, 2Urology, University of California-Los Angeles, Los Angeles, CA, United States
Prospectively undersampled 5D echo-planar J-resolved spectroscopic imaging (EP-JRESI) data were acquired in 9 prostate cancer patients and 3 healthy controls. The 5D data was reconstructed using Dictionary learning (DL), Total Variation (TV), Perona-Malik (PM) and a hybrid DLTV method combining DL and TV. DLTV uses the gradient sparsity of TV and the learned dictionary-based sparsity of DL to further increase the transform sparsity of the data. The DLTV method unambiguously resolved 2D J-resolved peaks including myo-inositol, citrate, creatine, spermine and choline with an improved reconstruction that facilitates higher acceleration factors, leading to significant reduction in scan time.
|
||
3486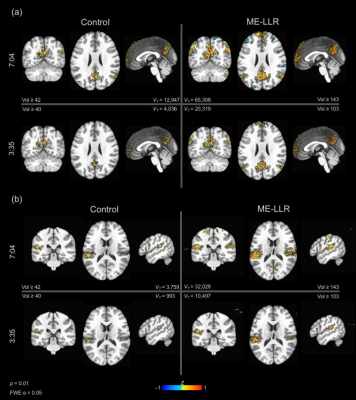 |
14 | Locally low-rank denoising of multi-echo fMRI data preserves detection of resting-state networks following scan truncation
Nolan K Meyer1,2, Daehun Kang2, Zaki Ahmed2, Myung-Ho In2, Yunhong Shu2, John Huston III2, Matt A Bernstein2, and Joshua D Trzasko2
1Mayo Clinic Graduate School of Biomedical Sciences, Rochester, MN, United States, 2Department of Radiology, Mayo Clinic, Rochester, MN, United States
Functional magnetic resonance imaging (fMRI) has inherent limitations of fast acquisitions due to low signal-to-noise ratio (SNR) and artifacts. Multi-echo fMRI acquires images at multiple TEs, increasing robustness to off-resonance based signal loss and improving sensitivity to neural activity. However, noise is substantial given limitations of EPI, although consistent across TEs contributing to prolonged scan times required for sufficient statistical power. Reduction of acquisition time with reconstruction and processing techniques is of interest. This study extends preliminary work on locally low-rank denoising of multi-echo fMRI data to explore scan time reduction through processing with retrospective truncation.
|
||
3487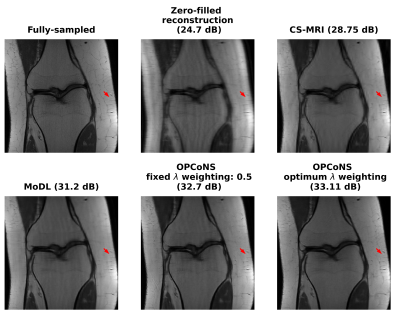 |
15 | Optimized Parallel Combination of Deep Networks and Sparsity Regularization for MR Image Reconstruction (OPCoNS)
Avrajit Ghosh1, Shijun Liang2, Anish Lahiri 3, and Saiprasad Ravishankar 1,2
1Department of Computational Mathematrics Science and Engineering, Michigan State University, East Lansing, MI, United States, 2Department of Biomedical Engineering, Michigan State University, East Lansing, MI, United States, 3Department of Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States
This work examines optimized parallel combinations of deep networks and conventional regularized reconstruction for improved quality of MR image reconstructions from undersampled k-space data. Features learned by deep networks and typical model-based iterative algorithms (e.g., sparsity-penalized reconstruction) could complement each other for effective reconstructions. We observe that combining the image features from multiple approaches in a parallel fashion with appropriate learned weights leads to more effective image representations that are not captured by either strictly supervised or (unsupervised) conventional iterative methods.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.