Online Gather.town Pitches
Machine Learning & Artificial Intelligence V
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
4803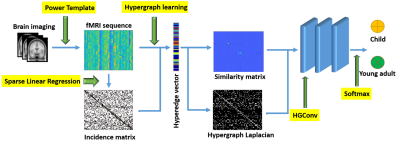 |
1 | Hypergraph learning-based convolutional neural network for classification of brain functional connectome
Junqi Wang1,2, Hailong Li1,2, Gang Qu3, Jonathan R Dillman1,2,4, Nehal A Parikh5,6, and Lili He1,2,4
1Department of Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 3Department of Biomedical Engineering, Tulane University, New Orleans, LA, United States, 4Department of Radiology, University of Cincinnati College of Medicine, Cincinnati, OH, United States, 5Center for Prevention of Neurodevelopmental Disorders, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 6Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
The human brain is a highly interconnected network where local activation patterns are organized to cope with diverse environmental demands. We developed a hypergraph learning based convolutional neural network model to capture higher order relationships between brain regions and learn representative features for brain connectome classification. The model was applied to a large scale resting state fMRI cohort, containing hundreds of healthy developing adolescents, age 8 to 22. The proposed model is able to classify different age groups with a balanced accuracy of 86.8%.
|
||
4804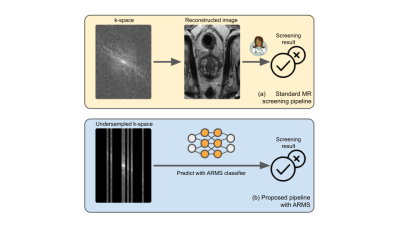 |
2 | Accelerated MR screenings with direct k-space classification
Raghav Singhal1, Mukund Sudarshan1, Luke Ginocchio2, Angela Tong2, Hersh Chandarana2, Daniel Sodickson2, Rajesh Ranganath3,4, and Sumit Chopra1,2
1Courant Institute of Mathematical Sciences, New York University, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research (CAI2R), Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 3Department of Population Health, New York University, New York, NY, United States, 4Center for Data Science, New York University, New York, NY, United States
Despite its rich clinical information content, MR has seen limited adoption in population-level screenings, due to concerns about specificity combined with high scan duration and cost. In order to begin to address such issues, and to accelerate the entire pipeline from data acquisition to diagnosis, we introduce ARMS, an algorithm that learns k-space undersampling patterns to maximize the accuracy of pathology detection. ARMS detects pathologies directly from undersampled k-space data, bypassing explicit image reconstruction. We use ARMS to detect clinically significant prostate cancer and knee abnormalities in 2D MR scans, achieving an acceleration of 12.5x without compromising accuracy.
|
||
4805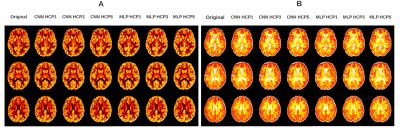 |
3 | Prediction of new diffusion MRI data is feasible using robust machine learning algorithms for multi-shell HARDI in a clinical setting
Cayden Murray1, Olayinka Oladosu1, and Yunyan Zhang 2,3
1Neuroscience, University of Calgary, Calgary, AB, Canada, 2Radiology, University of Calgary, Calgary, AB, Canada, 3Clinical Neurosciences, University of Calgary, Calgary, AB, Canada
High Angular Resolution Diffusion Imaging (HARDI) is a promising method for the analysis of microstructural changes. However, HARDI acquisition is time-consuming and therefore impractical in clinical settings. We developed 2 neural networks for predicting non-acquired diffusion datasets based on diffusion MRI: Multi-layer Perceptron (MLP) and Convolutional Neural Network (CNN). Through systemic training and evaluation with healthy public data and local MS patient MRI, we found that both the MLP and CNN models could predict high b-value from low b-value data that allowed the assessment of Neurite Orientation Dispersion and Density Imaging (NODDI) outcomes. Neural networks can make NODDI clinically viable.
|
||
 |
4806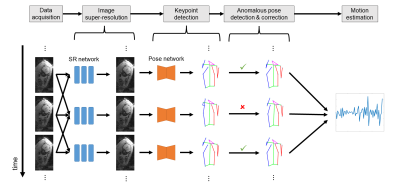 |
4 | An Automated Pose and Motion Estimation Pipeline in Dynamic 3D Fetal MRI
Junshen Xu1, Molin Zhang1, Lana Vasung2,3,4, Esra Abaci Turk2,3,4, Borjan Gagoski3,4,5, Polina Golland1,6, P. Ellen Grant2,3,4,5, and Elfar Adalsteinsson1,7
1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Department of Pediatrics, Boston Children’s Hospital, Boston, MA, United States, 3Harvard Medical School, Boston, MA, United States, 4Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 5Department of Radiology, Boston Children’s Hospital, Boston, MA, United States, 6Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 7Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Fetal motion is an important indicator of fetal health and nervous system development. Current assessments of fetal motion with MRI or ultrasound are qualitative and do not reflect the 3D motion of each body part . To study the detailed motion of fetuses, annotations of fetal pose are required, which would be time-consuming through manually-labelled data for each scan. In this work, we demonstrate an automated and efficient pipeline for fetal pose and motion estimation of fetal MRI using deep learning. The results of experiments show that the proposed pipeline outperforms other state-of-the-art fetal pose estimation methods.
|
|
4807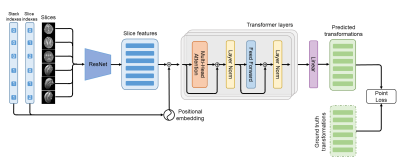 |
5 | SVoRT: Slice-to-volume Registration for Fetal Brain MRI Reconstruction with Transformers
Junshen Xu1, Daniel Moyer2, P. Ellen Grant3,4,5,6, Polina Golland1,2, Juan Eugenio Iglesias2,4,7,8, and Elfar Adalsteinsson1,9
1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 3Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4Harvard Medical School, Boston, MA, United States, 5Department of Pediatrics, Boston Children’s Hospital, Boston, MA, United States, 6Department of Radiology, Boston Children’s Hospital, Boston, MA, United States, 7Centre for Medical Image Computing, University College London, London, United Kingdom, 8Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Cambridge, MA, United States, 9Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Volumetric reconstruction of fetal brains from MR slices is a challenging task, which is sensitive to the initialization of slice-to-volume transformations. Further complicating the task is the unpredictable fetal motion. In this abstract, we proposed a novel method for slice-to-volume registration using transformers, which models the stacks of MR slices as a sequence. With the attention mechanism, the proposed model predicts the transformation of one slice using information from other slices. Results show that the proposed method achieves not only lower registration error but also better generalizability compared with other state-of-the-art methods for slice-to-volume registration of fetal MRI.
|
||
 |
4808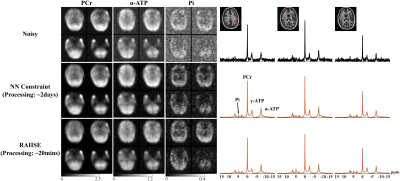 |
6 | LeaRning nonlineAr representatIon and projectIon for faSt constrained MRSI rEconstruction (RAIISE)
Yahang Li1,2, Loreen Ruhm3,4, Anke Henning3,4, and Fan Lam1,2,5
1Department of Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, Urbana, IL, United States, 3Advanced Imaging Research Center, University of Texas Southwestern Medical Center (UTSW), Dallas, TX, United States, 4Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 5Cancer Center at Illinois, Urbana, IL, United States We proposed here a novel method for computationally efficient reconstruction from noisy MRSI data. The proposed method is characterized by (a) a strategy that jointly learns a nonlinear low-dimensional representation of high-dimensional spectroscopic signals and a projector to recover the low-dimensional embeddings from noisy FIDs; and (b) a formulation that integrates forward encoding model, a spectral constraint from the learned representation and a complementary spatial constraint. The learned projector allows for the derivation of a highly efficient algorithm combining projected gradient descent and ADMM. The proposed method has been evaluated using simulation and in vivo data, demonstrating impressive SNR-enhancing performance. |
|
4809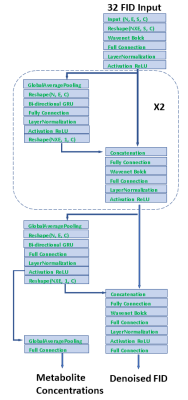 |
7 | A Deep Learning Neural Network for Quantifying Metabolite Concentrations by Multi-echo MRS
Yan Zhang1 and Jun Shen1
1National Institute of Mental Health, Bethesda, MD, United States
Multi echo techniques such as JPRESS consist of both short and long echoes and provide more diversified information for spectral fitting than techniques based on a single echo. However, fitting multi echo data is more challenging because signals attenuate with increasing echo time due to T2 relaxation, and the macromolecule background also varies across the echoes. We present a novel neural network architecture that directly maps the time domain JPRESS input onto metabolite concentrations. The testing results show the model can successfully predict in vivo metabolite concentrations from multi-echo JPRESS data after being trained with quantum mechanics simulated spectral data.
|
||
 |
4810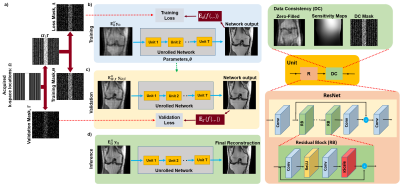 |
8 | Zero-Shot Physics-Guided Self-Supervised Learning for Subject-Specific MRI Reconstruction
Burhaneddin Yaman1,2, Seyed Amir Hossein Hosseini1,2, and Mehmet Akcakaya1,2
1Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, Minneapolis, MN, United States
While self-supervised learning enables training of deep learning reconstruction without fully-sampled data, it still requires a database. Moreover, performance of pretrained models may degrade when applied to out-of-distribution data. We propose a zero-shot subject-specific self-supervised learning via data undersampling (ZS-SSDU) method, where acquired data from a single scan is split into at least three disjoint sets, which are respectively used only in physics-guided neural network, to define training loss, and to establish an early stopping strategy to avoid overfitting. Results on knee and brain MRI show that ZS-SSDU achieves improved artifact-free reconstruction, while tackling generalization issues of trained database models.
|
|
4811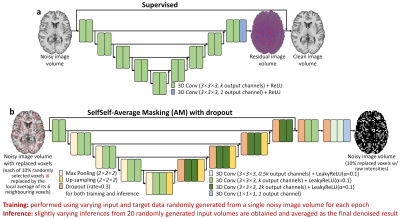 |
9 | Improving the accessibility of deep learning-based denoising for MRI using transfer learning and self-supervised learning
Qiyuan Tian1, Ziyu Li2, Wei-Ching Lo3, Berkin Bilgic1, Jonathan R. Polimeni1, and Susie Y. Huang1
1Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 2Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3Siemens Medical Solutions, Charlestown, MA, United States
The requirement for high-SNR reference data for training reduces the practical feasibility of supervised deep learning-based denoising. This study improves the accessibility of deep learning-based denoising for MRI using transfer learning that only requires high-SNR data of a single subject for fine-tuning a pre-trained convolutional neural network (CNN), or self-supervised learning that can train a CNN using only the noisy image volume itself. The effectiveness is demonstrated by denoising highly accelerated (R=3×3) Wave-CAIPI T1w MPRAGE images. Systematic and quantitative evaluation shows that deep learning without or with very limited high-SNR data can achieve high-quality image denoising and brain morphometry.
|
||
4812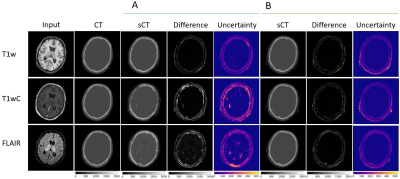 |
10 | Quantifying Domain Shift for Deep Learning Based Synthetic CT Generation with Uncertainty Predictions
Matt Hemsley1, Liam S.P Lawrence1, Rachel W Chan2, Brige Chuge3,4, and Angus Z Lau1,2
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Physical Sciences, Sunnybrook Research Insitute, Toronto, ON, Canada, 3Department of Radiation Oncology, Sunnybrook Health Sciences Centre, Toronto, ON, Canada, 4Department of Physics, Ryerson University, Toronto, ON, Canada
Convolutional Neural Networks behave unpredictively when test images differ from the training images, for example when different sequences or acquisition parameters are used. We trained models to generate synthetic CT images, and tested the models on both in-distribution and out-of-distribution input sequences to determine the magnitude of performance loss. Additionally, we evaluated if uncertainty estimates made using dropout-based variational inference could detect spatial regions of failure. Networks tested on out of distribution images failed to generate accurate synthetic CT images. Uncertainty estimates identified spatial regions of failure and increased with the difference between the training and testing sets.
|
||
4813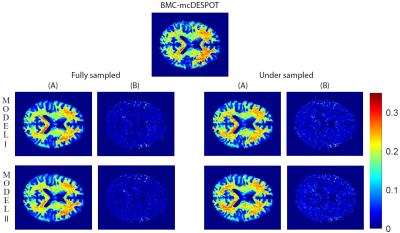 |
11 | Rapid myelin water fraction mapping through the combination of artificial neural network and under sampled mcDESPOT data
Zhaoyuan Gong1, Nikkita Khattar2, Matthew Kiely1, Curtis Triebswetter1, Maryam H. Alsameen1, and Mustapha Bouhrara1
1National Institute on Aging, Baltimore, MD, United States, 2Yale University, New Haven, CT, United States
The Myelin water fraction (MWF) measure provides a direct assessment of myelin content. The widely utilized method is the multicomponent analysis of T2 relaxation time and MWF is determined by the fraction of the fast-relaxing component. However, using either conventional or advanced methods, such as the BMC-mcDESPOT, requires prolonged acquisition and computation times, hampering their integration in clinical investigations. In this proof-of-concept work, we propose artificial neural network models to derive MWF maps from under sampled mcDESPOT data through two distinct approaches. This work opens the way to further developments for practical and rapid MWF imaging.
|
||
4814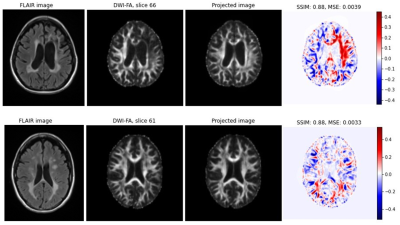 |
12 | Exploring the potential of StyleGAN projection for quantitative maps from diffusion-weighted MR images
Daniel Güllmar1, Wei-Chan Hsu1,2, Stefan Ropele3, and Jürgen R. Reichenbach1,2
1Institute of Diagnostic and Interventional Radiology, Medical Physics Group, Jena University Hospital, Jena, Germany, 2Michael Stifel Center Jena for Data-Driven and Simulation Science, Jena, Germany, 3Division of General Neurology, Medical University Graz, Graz, Austria
Synthetic medical images can be generated with a StyleGAN and are indistinguishable from real data even by experts. However, the projection of real data via latent space onto a synthetic image shows clear deviations from the original (at least on the second image). This plays a major role especially when using GANs to perform tasks such as image correction (e.g. noise reduction), image interpolation or image interpretation by analyzing the latent space. Based on the results shown, it is highly recommended to perform an analysis of the projection accuracy before applying any of these applications.
|
||
4815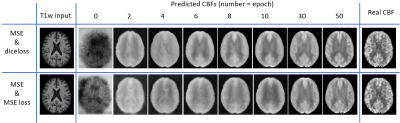 |
13 | A 3D-FiLM-cGAN Architecture for the Synthesis of Cerebral Blood Flow Maps
Michael Stritt1, Matthias Günther1,2,3, Johannes Gregori1,4, Daniel Mensing1, Henk-Jan Mutsaerts5, and Klaus Eickel1,3
1mediri GmbH, Heidelberg, Germany, 2Universität Bremen, Bremen, Germany, 3Fraunhofer MEVIS, Bremen, Germany, 4Darmstadt University of Applied Sciences, Darmstadt, Germany, 5Amsterdam University Medical Center, Amsterdam, Netherlands
The presented neural network with 3D-FiLM-cGAN architecture synthesizes cerebral blood flow maps from T1-weighted input images. Acquisition- and subject-specific metadata such as sex, arterial spin labeling (ASL) method and readout techniques were fed into the neural network as auxiliary input. The multi-vendor database including different ASL sequence types was created from ADNI data which were preprocessed in ExploreASL and transformed to MNI standard space. A subset of data from a single vendor (GE) were used for supervised training exemplarily and compared to CBF from acquired ASL data.
|
||
4816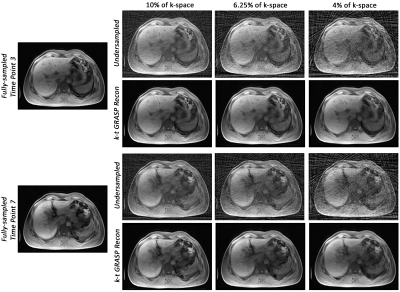 |
14 | Motion-robust dynamic abdominal MRI using k-t GRASP and dynamic dual-channel training of super-resolution U-Net (DDoS-UNet)
Chompunuch Sarasaen1,2,3, Soumick Chatterjee1,3,4,5, Georg Rose2,3, Andreas Nürnberger4,5,6, and Oliver Speck1,3,6,7,8
1Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Institute for Medical Engineering, Otto von Guericke University, Magdeburg, Germany, 3Research Campus STIMULATE, Otto von Guericke University, Magdeburg, Germany, 4Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 5Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Center for Behavioral Brain Sciences, Magdeburg, Germany, 8Leibniz Institute for Neurobiology, Magdeburg, Germany
Cartesian sampling techniques are available to speed up the measurement of dynamic MRI, such as k-t GRAPPA. However, radial samplings, such as iGRASP, are more robust to motion and can be applied for abdominal dynamic MRI. In this work, k-t GRAPPA inspired iGRASP has been created (so-called k-t GRASP)–which acquires the subsequent time points by starting the initial spoke of the time point with an angle with the last spoke of the previous time point, and extended by super-resolution reconstruction of dynamic abdominal MRI using DDoS-UNet. The method was evaluated in 3D dynamic data of four subjects with retrospective undersampling.
|
||
4817 |
15 | Improving Across-Dataset Brain Tissue Segmentation Using Transformer
Vishwanatha Mitnala Rao1, Zihan Wan2, David Ma1, Ye Tian1, and Jia Guo3
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Applied Mathematics, Columbia University, New York, NY, United States, 3Department of Psychiatry, Columbia University, New York, NY, United States Despite achieving compelling performance, many deep learning automated brain tissue segmentation solutions struggle to generalize to new datasets due to properties inherent to MRI scans. We propose TABS, a new transformer-based deep learning architecture that achieves state-of-the-art-performance, generalization, and consistency. We tested TABS on three datasets of differing field strands and acquisition parameters. TABS outperformed RAUnet on our performance testing and remained consistent across test-retest repeated scans from a separate dataset. Moreover, TABS achieved impressive generality performance and even improved in performance across datasets. We believe TABS represents a generalized and accurate brain tissue segmentation alternative. |
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.