Online Gather.town Pitches
Machine Learning & Artificial Intelligence II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
3458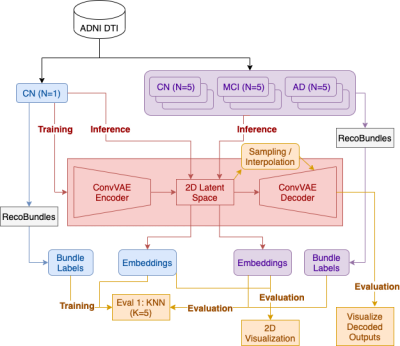 |
1 | Deep generative model for learning tractography streamline embeddings based on a Convolutional Variational Autoencoder
Yixue Feng1, Bramsh Qamar Chandio1,2, Tamoghna Chattopadhyay1, Sophia I. Thomopoulos1, Conor Owens-Walton1, Neda Jahanshad1, Eleftherios Garyfallidis2, and Paul M. Thompson1
1Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Marina Del Rey, CA, United States, 2Department of Intelligent Systems Engineering, School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, IN, United States
We present a deep generative model to autoencode tractography streamlines into a smooth low dimensional latent distribution, which captures their spatial and sequential information with 1D convolutional layers. Using linear interpolation, we show that the learned latent space translates smoothly into the streamline space and can decode meaningful outputs from sampled points. This allows for inference on new data and direct use of Euclidean distance on the embeddings for downstream tasks, such as bundle labeling, quantitative inter-subject comparisons, and group statistics.
|
||
3459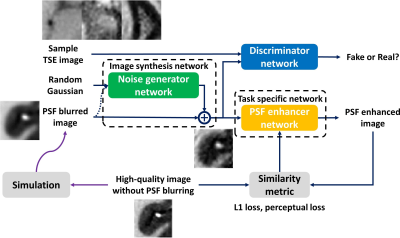 |
2 | Unsupervised Domain Adaptation for Neural Network Enhanced Turbo Spin Echo Imaging
Zechen Zhou1, Peter Börnert2, and Chun Yuan3
1Philips Research North America, Seattle, WA, United States, 2Philips Research Hamburg, Hamburg, Germany, 3Vascular Imaging Lab, University of Washington, Seattle, WA, United States
Supervised learning is widely used for deep learning based image quality enhancement for improved clinical diagnosis. However, the difficulties to acquire a large number of high-quality reference image for different MR applications can limit its generalization performance. An unsupervised domain adaptation (DA) approach is proposed and incorporated into the deep learning based image enhancement framework, which improves the performance of trained network on new datasets. Preliminary evaluation on point spread function enhanced turbo spin echo imaging has showed that the unsupervised DA approach can provide more stabilized image sharpness improvement without severe amplified noise.
|
||
3460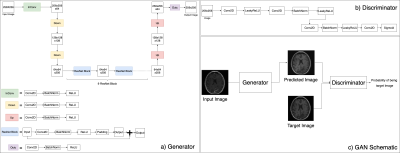 |
3 | Using deep learning to generate missing anatomical imaging contrasts required for lesion segmentation in patients with glioma.
Ozan Genc1, Pranathi Chunduru2, Annette Molinaro1, Valentina Pedoia1, Susan Chang1, Javier Villanueva-Meyer1, and Janine Lupo Palladino1
1University of California San Francisco, San Francisco, CA, United States, 2Johnson & Johnson, San Francisco, CA, United States
Missing value imputation is an important concept in statistical analyses. We utilized conditional GAN based deep learning models to learn missing contrasts in MR images. We trained two deep learning models (FSE to FLAIR and T1 post GAD to T1 pre-GAD) for MR image conversion for missing value imputation. The model performances are evaluated by visual examination and comparing SSIM values. We observed that these models can learn the output contrast.
|
||
3461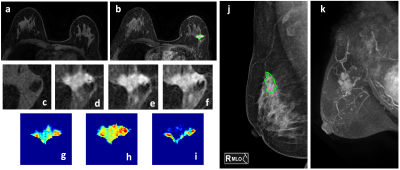 |
4 | Mammography Lesion ROI Drawing Guided by Breast MRI MIP to Extract Features from Corresponding Lesions to Build Radiomics Diagnostic Models
Yan-Lin Liu1, Zhongwei Chen2, Youfan Zhao2, Yang Zhang1,3, Jiejie Zhou2, Jeon-Hor Chen1, Ke Nie3, Meihao Wang2, and Min-Ying Su1
1Department of Radiological Sciences, University of California, Irvine, CA, United States, 2Department of Radiology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China, 3Department of Radiation Oncology, Rutgers-Cancer Institute of New Jersey, Robert Wood Johnson Medical School, New Brunswick, NJ, United States
268 patients with DCE-MRI and mammography were analyzed to evaluate the diagnostic performance of radiomics models. The dataset was split to 202 (146 malignant 56 benign) for training, and 66 (48 malignant 18 benign) for testing. The MIP of MR contrast enhancement maps was generated to simulate the CC and MLO view as guidance for manual ROI drawing on mammography. The models were built using features extracted by PyRadiomics. Combined MRI and mammography features can reach 89.6% accuracy in training and 83.3% in testing datasets, and the addition of mammography can improve specificity while maintaining high sensitivity of MRI.
|
||
3462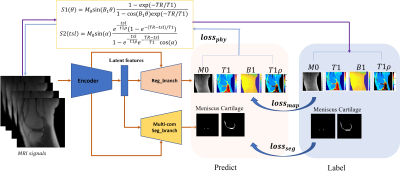 |
5 | Deep CNNs with Physical Constraints for simultaneous Multi-tissue Segmentation and Multi-parameter Quantification (MSMQ-Net) of Knee
Xing Lu1, Yajun Ma1, Kody Xu1, Saeed Jerban1, Hyungseok Jang1, Chun-Nan Hsu2, Amilcare Gentili1,3, Eric Y Chang1,3, and Jiang Du1
1Department of Radiology, University of California, San Diego, San Diego, CA, United States, 2Department of Neurosciences, University of California, San Diego, San Diego, CA, United States, 3Radiology Service, Veterans Affairs San Diego Healthcare System, San Diego, CA, United States
In this study, we proposed end-to-end deep learning convolutional neural networks to perform simultaneous multi-tissue segmentation and multi-parameter quantification (MSMQ-Net) on the knee without and with physical constraints. The performance robustness of MSMQ-Net was also evaluated using reduced input magnetic resonance images. Results demonstrated the potential of MSMQ-Net for fast and accurate UTE-MRI analysis of the knee, a “whole-organ” approach which is impossible with conventional clinical MRI.
|
||
3463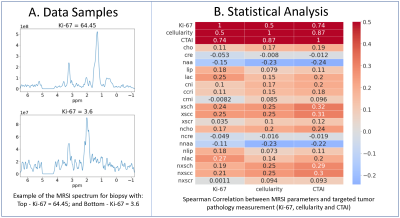 |
6 | 1H-MRS and machine learning for predicting voxel-wise histopathology of tumor cells in newly-diagnosed glioma patients
Nate Tran1, Jacob Ellison1, Oluwaseun Adegbite1, James Golden1, Yan Li1, Joanna Phillips2, Devika Nair1, Anny Shai2, Annette Molinaro2, Valentina Pedoia1, Javier Villanueva-Meyer1, Mitchel Berger2, Shawn Hervey-Jumper2, Aghi Manish2, Susan Chang2, and Janine Lupo1
1Radiology & Biomedical Imaging, University of California, San Francisco, SAN FRANCISCO, CA, United States, 2Neurological Surgery, University of California, San Francisco, SAN FRANCISCO, CA, United States
Using spectrum obtained at the spatial location of 549 tissue samples from 261 newly diagnosed patients with glioma, we trained and tested an support vector regression (SVR) model on individual metabolites, and a 1D-CNN model on the whole spectrum, to predict tumor biology such as cellularity, Ki-67, and tumor aggressiveness. A regression based 1D-CNN model using the entire spectrum pre-trained on a similar classification task outperformed the SVR model using metabolite peak heights.
|
||
3464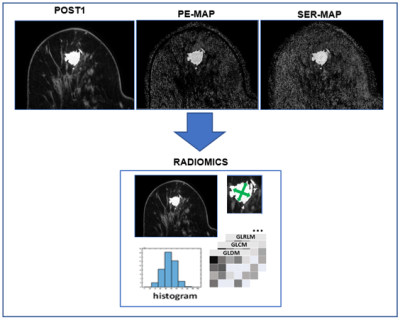 |
7 | Radiomics to Predict Pathological Complete Response in Patients with Triple Negative Breast Cancer
Michael Hirano1, Anum S. Kazerouni2, Mladen Zecevic2, Laura C. Kennedy3, Shaveta Vinayak2, Habib Rahbar2, Matthew J. Nyflot2, Suzanne Dintzis2, and Savannah C. Partridge2
1University of Washingon, Seattle, WA, United States, 2University of Washington, Seattle, WA, United States, 3Vanderbilt University, Nashville, TN, United States
Radiomics is an advancing field of medical image analysis based on extracting large sets of quantitative features that can be used for outcome modeling for clinical decision support. Our study investigated the value of radiomics features extracted from pre-treatment dynamic contrast-enhanced MRI for the prediction of neoadjuvant chemotherapy response in patients with triple-negative breast cancer. In a retrospective cohort of 103 TNBC patients, radiomics-based models using post-contrast images and kinetics maps were moderately predictive of pathologic response, and lesion size and shape features were the most consistent predictors across all image types.
|
||
3465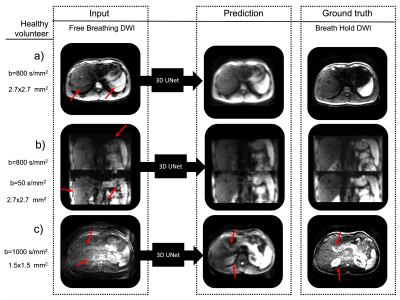 |
8 | Predicting Breath-Hold Liver Diffusion MRI from Free-Breathing Data using a Convolutional Neural Network (CNN) Video Permission Withheld
Emmanuelle M. M. Weber1, Xucheng Zhu2, Patrick Koon2, Anja Brau2, Shreyas Vasanawala1, and Jennifer A. McNab1
1Stanford, Stanford, CA, United States, 2GE Healthcare, Menlo Park, CA, United States
To reduce artifacts in free breathing single-shot diffusion MRI of the liver, UNet based convolutional neural networks were trained to predict breath-hold data from free-breathing data using: 1) simulated data based on a digital phantom and 2) 31 scans of a healthy volunteer. The developed networks successfully reduced motion induced artifacts in DWI images.
|
||
 |
3466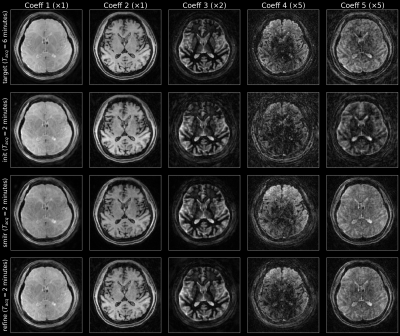 |
9 | SMILR - Subspace MachIne Learning Reconstruction
Siddharth Srinivasan Iyer1,2, Christopher M. Sandino3, Mahmut Yurt3, Xiaozhi Cao2, Congyu Liao2, Sophie Schauman2, and Kawin Setsompop2,3
1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States Recent developments in spatiotemporal MRI techniques enable whole-brain multi-parametric mapping in incredibly short acquisition times through highly-efficient k-space encoding, subspace reconstruction and carefully-designed regularization. However, this comes at the cost of long reconstruction times making such methods difficult to integrate into clinical practice. This abstract proposes a framework denoted SMILR (pronounced smile-r) to reduce the reconstruction times of subspace methods from multiple hours to a few minutes through machine learning. To evaluate performance, the framework is applied to multi-axis spiral projection MRF (denoted SPI-MRF) where it achieves improved reconstruction over conventional subspace reconstruction with locally low-rank at ~16-20x faster speed. |
|
3467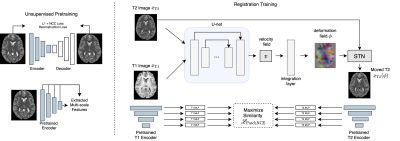 |
10 | Contrastive Learning of Inter-domain Similarity for Unsupervised Multi-modality Deformable Registration
Neel Dey1, Jo Schlemper2, Seyed Sadegh Mohseni Salehi2, Bo Zhou3, and Michal Sofka2
1Computer Science and Engineering, New York University, New York City, NY, United States, 2Hyperfine, New York City, NY, United States, 3Yale University, New Haven, CT, United States
We propose an unsupervised contrastive representation learning framework for deformable and diffeomorphic multi-modality MR image registration. The proposed deep network and data-driven objective function yield improved registration performance in terms of anatomical volume overlap over several previous hand-crafted objectives such as Mutual Information and others. For fair comparison, our experiments train all methods over the entire range of a key registration hyperparameter controlling deformation smoothness using conditional registration hypernetworks. T1w and T2w brain MRI registration improvements are presented across a large cohort of 1041 high-field 3T research-grade acquisitions while maintaining comparable deformation smoothness and invertibility characteristics to previous methods.
|
||
3468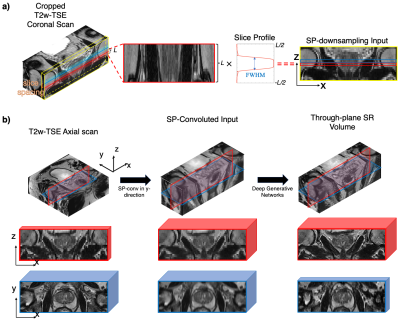 |
11 | Super-resolution MRI using Novel Slice-profile Based Transformation for Multi-slice 2D TSE Imaging
Jiahao Lin1,2, Miao Qi1,3, Chuthaporn Surawech1,4,5, Steven S Raman1, Holden Wu1, and Kyunghyun Sung1
1Department of Radiology, University of California, Los Angeles, Los Angeles, CA, United States, 2Department of Electrical and Computer Engineering, University of California, Los Angeles, Los Angeles, CA, United States, 3Department of Radiology, The First Affiliated Hospital of China Medical University, Shenyang, China, 4Department of Radiology, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, 5Division of Diagnostic Radiology, Department of Radiology, King Chulalongkorn Memorial Hospital, Bangkok, Thailand
Multi-slice two-dimensional turbo spin-echo (2D TSE) imaging is commonly used for its excellent in-plane resolution. However, multiple 2D TSE scans are acquired due to their poor through-plane resolution. In this study, we propose a novel slice-profile-based transformation super-resolution (SPTSR) framework for through-plane super-resolution (SR) of multi-slice 2D TSE imaging. We utilized a large clinical MRI dataset in this study. We demonstrate the effectiveness of our proposed SPTSR framework in 5.5x through-plane SR by both the visual comparison results and reader study results.
|
||
3469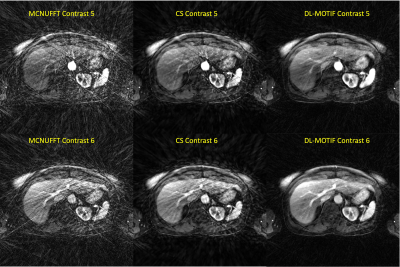 |
12 | DL-MOTIF: Deep Learning Based Motion Transformation Integrated Forward-Fourier Reconstruction for Free-Breathing Liver DCE-MRI
Sihao Chen1, Weijie Gan1, Cihat Eldeniz1, Ulugbek S. Kamilov1, Tyler J. Fraum1, and Hongyu An1
1Washington University in St. Louis, Saint Louis, MO, United States
Dynamic contrast-enhanced MRI (DCE-MRI) of the liver offers structural and functional information for assessing the contrast uptake visually. However, respiratory motion and the requirement of high temporal resolution make it difficult to generate high-quality DCE-MRI. In this study, we proposed a novel deep learning based motion transformation integrated forward-Fourier (DL-MOTIF) reconstruction using motion fields derived from a deep learning Phase2Phase (P2P) network and deep learning priors from a residual network on severely undersampled DCE. This approach reconstructs sharp motion-free DCE images with artifacts removal by incorporating deep learning motion fields for motion integration and deep learning priors for regularization.
|
||
3470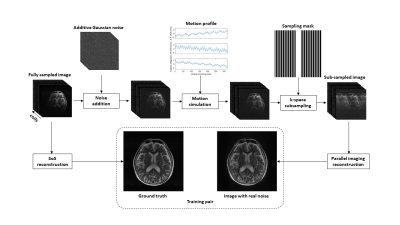 |
13 | Deep learning-based brain MRI reconstruction with realistic noise
Quan Dou1, Xue Feng1, and Craig H. Meyer1
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States
Fast imaging techniques can speed up MRI acquisition but can also be corrupted by noise, reconstruction artifacts, and motion artifacts in a clinical setting. A deep learning-based method was developed to reduce imaging noise and artifacts. A network trained with the supervised approach improved the image quality for both simulated and in vivo data.
|
||
3471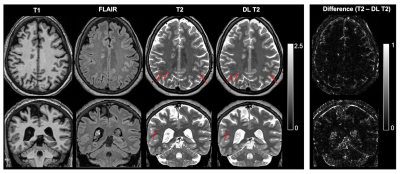 |
14 | Perivascular Space Quantification with Deep Learning synthesized T2 from T1w and FLAIR images
Jiehua Li1, Pan Su1,2, Rao Gullapalli1, and Jiachen Zhuo1
1Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 2Siemens Medical Solutions USA, Inc., Malvern, PA, United States
The perivascular space (PVS) plays a major role in brain waste clearance and brain metabolic homeostasis. Enlarged PVS (ePVS) is associated with many neurological disorders. ePVS is best depicted as hyper-intensities T2w images and can be reliable quantified with both the 3D T1w and T2w images. However many studies opt to acquire 3D FLAIR images instead of T2w due to its high specificity to white matter abnormalities (e.g. the ADNI study). Here we show that deep learning techniques can be used to synthesize T2w images from T1w & FLAIR images and improve the ePVS quantification in absence of T2w images.
|
||
3472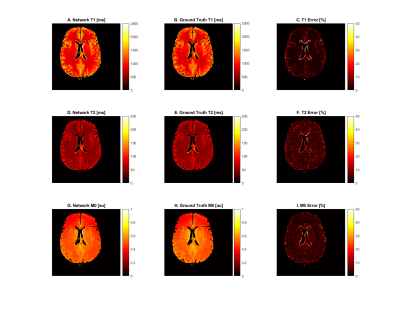 |
15 | Self-supervised and physics informed deep learning model for prediction of multiple tissue parameters from MR Fingerprinting data
Richard James Adams1, Yong Chen1, Pew-Thian Yap2, and Dan Ma1
1Case Western Reserve University, Cleveland, OH, United States, 2University of North Carolina, Chapel Hill, NC, United States
Magnetic resonance fingerprinting (MRF) simultaneously quantifies multiple tissue properties. Deep learning accelerates MRF’s tissue mapping time; however, previous deep learning MRF models are supervised, requiring ground truths of tissue property maps. It is challenging to acquire quality reference maps, especially as the number of tissue parameters increases. We propose a self-supervised model informed by the physical model of the MRF acquisition without requiring ground truth references. Additionally, we construct a forward model that directly estimates the gradients of the Bloch equations. This approach is flexible for modeling MRF sequences with pseudo-randomized sequence designs where an analytical model is not available.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.