Oral Session
Deep/Machine Learning-Based Quantitative Imaging & Modeling
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

09:15 |
0433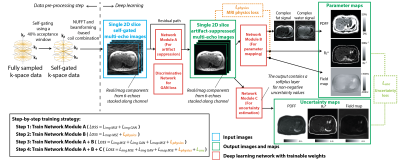 |
Uncertainty-Aware Physics-Driven Deep Learning Network for Fat and R2* Quantification in Self-Gated Free-Breathing Stack-of-Radial MRI
Shu-Fu Shih1,2, Sevgi Gokce Kafali1,2, Kara L. Calkins3, and Holden H. Wu1,2
1Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, United States, 2Department of Bioengineering, University of California Los Angeles, Los Angeles, CA, United States, 3Department of Pediatrics, University of California Los Angeles, Los Angeles, CA, United States
MRI noninvasively quantifies liver fat and iron in terms of proton-density fat fraction (PDFF) and R2*. While conventional Cartesian-based methods require breath-holding, recent self-gated free-breathing radial techniques have shown accurate and repeatable PDFF and R2* mapping. However, data oversampling or computationally expensive reconstruction is required to reduce radial undersampling artifacts due to self-gating. This work developed an uncertainty-aware physics-driven deep learning network (UP-Net) that accurately and rapidly quantifies PDFF and R2* using data from self-gated free-breathing stack-of-radial MRI. UP-Net used an MRI physics loss term to guide quantitative mapping, and also provided uncertainty estimation for each quantitative parameter.
|
|
| 09:27 | 0434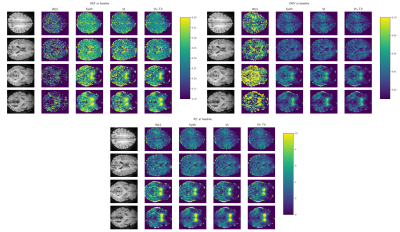 |
Improving oxygenation quantification from streamlined qBOLD data using amortized variational inference
Ivor J. A. Simpson1, Ashley McManamon 1, Alan J Stone2, Nicholas P Blockley3, Alessandro Colasanti4, and Mara Cercignani5
1Department of Informatics, University of Sussex, Brighton, United Kingdom, 2Department of Medical Physics and Clinical Engineering, St. Vincent's University Hospital, Dublin, Ireland, 3School of Life Sciences, University of Nottingham, Nottingham, United Kingdom, 4Brighton and Sussex Medical School, Brighton, United Kingdom, 5CUBRIC, Cardiff University, Cardiff, United Kingdom Streamlined qBOLD acquisitions enable experimentally straightforward observations of brain metabolism. High quality R2’ maps are easily derived; however, the oxygen extraction fraction (OEF) and deoxygenated blood volume (DBV) are more ambiguously defined from noisy data. Accordingly, standard approaches yield noisy and underestimated OEF maps and overestimate DBV. This work uses synthetic data to learn models for voxelwise prior distributions, which are subsequently leveraged in an amortized variational Bayesian inference model. We demonstrate our approach enables inference of smooth OEF and DBV maps, with a physiologically realistic distribution, and illustrate voxelwise differences in OEF between subjects at rest and undergoing hyperventilations. |
|
| 09:39 | 0435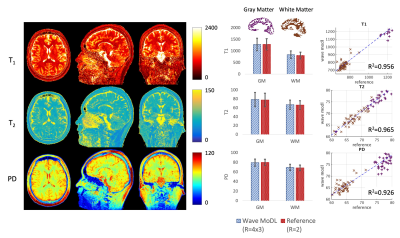 |
Rapid Quantitative Imaging Using Wave-Encoded Model-Based Deep Learning for Joint Reconstruction
Jaejin Cho1,2, Borjan Gagoski2,3, Tae Hyung Kim1,2, Qiyuan Tian1,2, Robert Frost1,2, Itthi Chatnuntawech4, and Berkin Bilgic1,2,5
1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4National Nanotechnology Center, Pathum Thani, Thailand, 5Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
We propose a wave-encoded model-based deep learning (wave-MoDL) method for joint multi-contrast image reconstruction with volumetric encoding using an interleaved look-locker acquisition sequence with T2 preparation pulse (3D-QALAS). Wave-MoDL enables a 2-minute acquisition at R=4x3-fold acceleration using a 32-channel array to provide T1, T2, and proton density maps at 1 mm isotropic resolution, from which standard contrast-weighted images can also be synthesized.
|
|
| 09:51 | 0436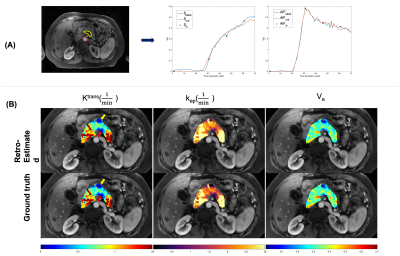 |
Retrospective Pharmacokinetic Quantification of Clinical Abdominal DCE-MRI using Deep Learning
Chaowei Wu1,2, Nan Wang3, Srinivas Gaddam4, Hui Han1, Stephen Pandol4, Anthony G. Christodoulou1,2, Yibin Xie1, and Debiao Li1,2
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 3Radiology Department, Stanford University, Stanford, CA, United States, 4Division of Digestive and Liver Diseases, Cedars-Sinai Medical Center, Los Angeles, CA, United States Quantitative dynamic contrast-enhanced (DCE) MRI has the potential for early detection, accurate staging, and therapy monitoring of cancers. However, clinical abdominal DCE-MRI has limited temporal resolution and can only provide qualitative or semi-quantitative assessments of tissue vascularity. In this study, we investigated the feasibility of retrospective quantification of multi-phasic abdominal DCE-MRI by improving the temporal resolution via deep learning. Simulated multi-phasic DCE data was generated using 2-sec temporal-resolution Multitasking DCE images. Results show that DCE kinetic parameters retrospectively estimated by deep learning agree with the ground truth, and are capable of differentiating abnormal tissues. |
|
| 10:03 | 0437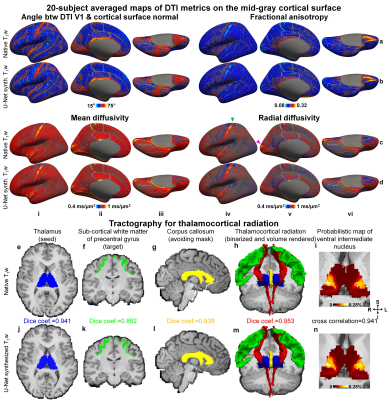 |
Diffusion MRI data analysis using brain segmentation from anatomical images synthesized from diffusion data by deep learning (DeepAnat)
Ziyu Li1, Qiuyun Fan2,3, Berkin Bilgic2,3, Guangzhi Wang4, Jonathan R Polimeni2,3, Susie Y Huang2,3, and Qiyuan Tian2,3
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 3Harvard Medical School, Boston, MA, United States, 4Department of Biomedical Engineering, Tsinghua University, Beijing, China
The analysis of diffusion MRI data requires brain segmentation from separate anatomical images, which may be unavailable or cannot be accurately co-registered to diffusion images due to image distortions in diffusion data. Two state-of-the-art convolutional neural networks, U-Net and generative adversarial network (GAN), are employed to synthesize high-quality, distortion-matched T1w images directly from diffusion data, suitable for generating accurate cerebral cortical surfaces and volumetric segmentation for surface-based analysis of DTI metrics and tractography. The accuracy is quantitatively evaluated, and the systematical comparison shows that GAN-synthesized images are more visually appealing while U-Net-synthesized images achieve higher data consistency and segmentation accuracy.
|
|
| 10:15 | 0438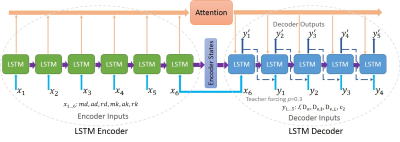 |
Direct parameter estimation of white matter model from DKI maps using recurrent neural network
Yujian Diao1,2,3 and Ileana Ozana Jelescu2,4
1Laboratory of Functional and Metabolic Imaging, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 3Animal Imaging and Technology, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 4Department of Radiology, Lausanne University Hospital, Lausanne, Switzerland
WMTI-Watson is a widely used biophysical model that estimates microstructure parameters from the diffusion and kurtosis tensors. Here we propose a deep learning (DL) approach based on the recurrent neural network (RNN) to increase the robustness and accelerate the parameter estimation. The RNN solver achieved high accuracy, had good generality and was extremely fast in computation. The proposed DL approach is highly promising to replace the conventional nonlinear least-squares optimization in parameter estimation of WMTI-Watson model and thus estimate WM parameters from any DKI maps.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.