Power Pitch
Pitch: Quantitative Relaxation Parameter Mapping: Accuracy, Robustness & Analysis
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

Power Pitch Session: How it Works
1st Hour: 2-minute Power Pitches in the Power Pitch Theater.
2nd Hour: 60-minute digital poster presentations at the smaller screens around the perimeter of the Power Pitch Theater.
16:45 |
0365.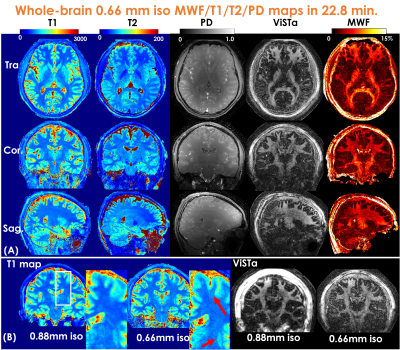 |
Mesoscale myelin-water fraction and T1/T2/PD mapping through optimized 3D ViSTa-MRF and stochastic reconstruction with preconditioning
Congyu Liao1,2, Xiaozhi Cao1,2, Siddharth Srinivasan Iyer1,3, Zihan Zhou4, Yunsong Liu5, Justin Haldar5, Mahmut Yurt1,2, Ting Gong6, Zhe Wu7, Hongjian He4, Jianhui Zhong4,8, Adam Kerr1,9, and Kawin Setsompop1,2
1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 4Center for Brain Imaging Science and Technology, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 5Signal and Image Processing Institute, University of Southern California, Los Angeles, CA, United States, 6Centre for Medical Imaging Computing, Department of Computer Science, University College London, London, United Kingdom, 7Techna Institute, University Health Network, Toronto, ON, Canada, 8Department of Imaging Sciences, University of Rochester, Rochester, NY, United States, 9Stanford Center for Cognitive and Neurobiological Imaging, Stanford University, Stanford, CA, United States
In this work, we developed ViSTa-MRF, which combined Visualization of Short Transverse relaxation time component (ViSTa) technique with MR Fingerprinting (MRF), to achieve high-fidelity whole-brain myelin-water fraction (MWF) and T1/T2/PD mapping at sub-millimeter isotropic resolution on a clinical 3T scanner. To achieve fast acquisition and memory-efficient reconstruction, the ViSTa-MRF sequence leverages an optimized 3D tiny-golden-angle-shuffling (TGAS) spiral-projection acquisition and stochastic subspace reconstruction with optimized k-space diagonal preconditioning. With the proposed ViSTa-MRF method, high-fidelity direct MWF mapping was achieved without a need for multi-compartment fitting.
|
|
16:47 |
0366.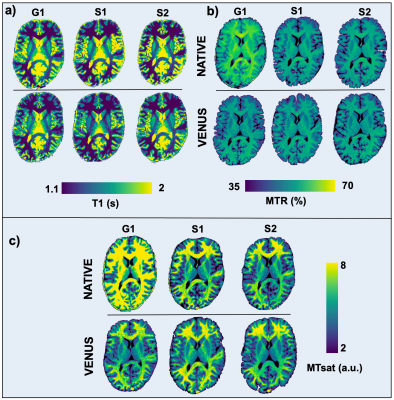 |
Multicenter reproducibility of quantitative MRI using vendor-neutral sequences (VENUS)
Agah Karakuzu1,2, Julien Cohen-Adad1,3, and Nikola Stikov1,2
1NeuroPoly Lab, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Heart Institute, Montreal, QC, Canada, 3Unité de Neuroimagerie Fonctionnelle (UNF), Centre de recherche de l’Institut Universitaire de Gériatrie de Montréal (CRIUGM), Montreal, QC, Canada
Vendor-native implementations can lead to unaccounted variance in quantitative MRI (qMRI). The purpose of this study is to show that fully transparent qMRI workflows coupled with VEndor-NeUtral Sequences (VENUS) improve multicenter reproducibility of quantitative MRI maps across three scanners by two vendors, in phantoms and in-vivo. We developed and deployed a vendor-neutral 3D spoiled gradient-echo (SPGR) sequence on three commercial scanners and showed that vendor-neutral qMRI maps of T1, MTR and MTsat outperform their vendor-native counterparts in terms of reproducibility.
|
|
16:49 |
0367.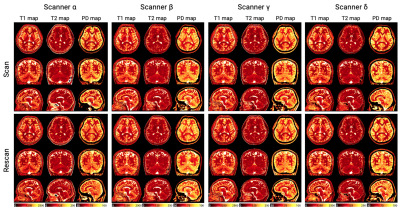 |
Multi-vendor comparison of three-dimensional multi-parametric mapping of the human brain at 3T: A multi-center study
Shohei Fujita1,2, Borjan Gagoski3,4, Ken-Pin Hwang5, Marcel Warntjes6,7, Akifumi Hagiwara1, Issei Fukunaga1, Wataru Uchida1, Yuma Nishimura1, Tomoya Muroi1, Toshiya Akatsu1, Akihiro Kasahara2, Ryo Sato2, Tsuyoshi Ueyama2, Christina Andica1, Koji Kamagata1, Shiori Amemiya2, Hidemasa Takao2, Osamu Abe2, and Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Fetal Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Imaging Physics, The University of Texas MD Anderson Cancer Center, Houston, TX, United States, 6SyntheticMR, Linköping, Sweden, 7Center for Medical Imaging Science and Visualization (CMIV), Linköping University, Linköping, Sweden Cross-vendor techniques enabling the pooling of data among different vendors is desired in multi-center projects; however, few mapping techniques have shown compatibility. In this study, we examined the intra-scanner repeatability and inter-vendor reproducibility of quantitative measurements obtained via a multi-parametric mapping technique, 3D-QALAS. Volunteers underwent scan-rescan sessions on four different 3T systems from three MRI vendors (GE, Philips, and Siemens). T1, T2, and PD values and brain tissue volumes of healthy volunteers derived from 3D-QALAS were robust across scanners from different vendors (inter-vendor coefficient of variation=3.3%). The data in this study will inform future technical developments for 3D-QALAS. |
|
| 16:51 | 0368.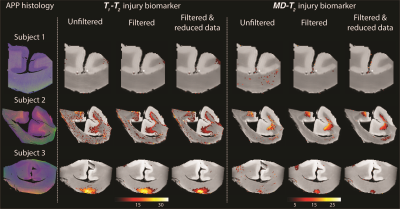 |
Microscopic neuropathology can be seen using accelerated combined diffusion-relaxation MRI
Dan Benjamini1, Mustapha Bouhrara1, Michal E Komlosh2, Diego Iacono3, Daniel P Perl3, David L Brody3, and Peter J Basser2
1National Institute on Aging, Baltimore, MD, United States, 2National Institute of Child Health and Human Development, Bethesda, MD, United States, 3Uniformed Services University of the Health Sciences, Bethesda, MD, United States Here we present MRI acquisitions that maximize the amount of information in an image by using a combination of magnetic field profiles to probe relaxation and diffusion mechanisms simultaneously, thus resolving sub-voxel information from ex vivo brain segments of human subjects that sustained traumatic brain injury. We then propose a strategy to refine the ensuing abundance of high-dimensional data, and to sort and recover biologically relevant information into actionable and quantitative injury biomarker images that capture subtle neuropathology. |
|
16:53 |
0369.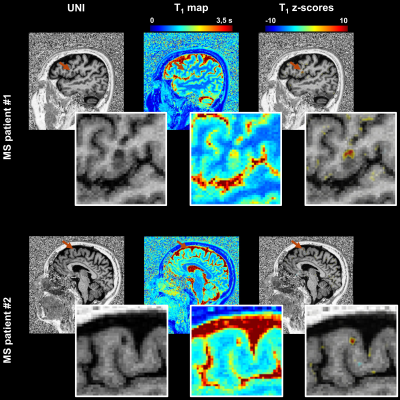 |
Quantitative atlas of T1 relaxation times in the cerebral cortex for personalized characterization of pathology
Gian Franco Piredda1,2,3, Marion Claudet1, Tom Hilbert1,2,3, Manuela Vaneckova4, Jan Krasensky4, Michaela Andelova5, Tomas Uher5, Eva Kubala Havrdova5, Karolina Vodehnalova5, Dana Horakova5, Karl Egger6, Shan Yang6, Riccardo Ludovichetti7, Lindsey A. Crowe7, Bénédicte M. A. Delattre7, Maria Isabel Vargas7, Veronica Ravano1,2,3, Bénédicte Maréchal1,2,3, Jean-Philippe Thiran2,3, and Tobias Kober1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4Department of Radiology, First Faculty of Medicine, Charles University and General University Hospital, Prague, Czech Republic, 5Department of Neurology and Center of Clinical Neuroscience, First Faculty of Medicine, Charles University and General University Hospital, Prague, Czech Republic, 6Department of Neuroradiology, Medical Center – University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany, 7Divsion of Neuroradiology, Department of Radiology and Medical Informatics, Hôpitaux Universitaires de Genève (HUG), Geneva, Switzerland
Comparing single-patient data to atlases of normative relaxation times in the brain enables personalized characterization of pathological tissue. However, an accurate comparison is challenging in the cortex due to the gyrification of gray matter and resulting registration problems. This work addresses this problem for parametric T1 maps that were acquired from 285 healthy subjects. A method aligning inter-subject brain cortices was developed to enable the comparison of cortical T1 values. We thus established a normative atlas accounting for healthy T1 values in brain cortical tissues, to be used to detect and characterize pathology-induced T1 alterations in patients.
|
|
16:55 |
0370.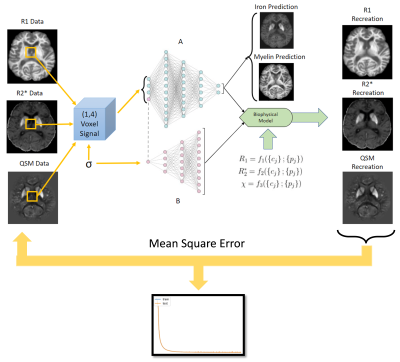 |
Beyond qMRI: Biological tissue properties from single-subject unsupervised deep learning with theoretical signal constraints
Ilyes Benslimane1, Günther Grabner2, Simon Hametner3, Thomas Jochmann1,4, Robert Zivadinov1,5, and Ferdinand Schweser1,5
1Department of Neurology, Buffalo Neuroimaging Analysis Center, Buffalo, NY, United States, 2Department of Medical Engineering, Carinthia University of Applied Sciences, Klagenfurt, Austria, 3Department of Biomedical Imaging and Image-Guided Therapy, High Field MR Centre, Medical University of Vienna, Austria, 4Department of Computer Science and Automation, Technische Universität Ilmenau, Ilmenau, Germany, 5Center for Biomedical Imaging, Clinical and Translational Science Institute at the University at Buffalo, Buffalo, NY, United States
Most quantitative MRI (qMRI) parameters co-depend on multiple tissue properties, limiting their clinical value. Here, we introduce a biophysically constrained autoencoder network that accepts multiparametric MR data and outputs underlying biological tissue parameters. The method requires no ground truth data, no knowledge of the (generally unknown) biophysical model parameters, and a single subject provides sufficient training data. We validated the method using ex vivo iron and myelin stains.
|
|
16:57 |
0371.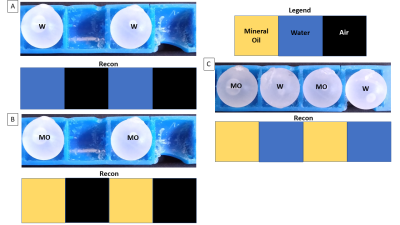 |
Selective Encoding through Nutation and Fingerprinting (SENF): 1D Experimental Validation on a 47.5 mT Low-Field Scanner
Christopher E Vaughn1,2, N Reid Bolding3, Mark A Griswold4, and William A Grissom1,2
1Department of Biomedical Engineering, Vanderbilt University, Nashville, TN, United States, 2VUIIS, Vanderbilt University, Nashville, TN, United States, 3Physics, Case Western Reserve University, Cleveland, OH, United States, 4Radiology, Case Western Reserve University, Cleveland, OH, United States
B0 gradients have several problems including cost, bulkiness, noise, and PNS. We propose to use a method called Selective Encoding through Nutation and Fingerprinting (SENF) which is a RF spatial encoding technique that also encodes quantitative information. In this abstract we validate SENF on a 47.5 mT low-field scanner using a high-flip MR Fingerprinting type sequence with a gradient RF coil to perform a 4-voxel 1D reconstruction that can differentiate between three materials (mineral oil, water, and air).
|
|
| 16:59 | 0372.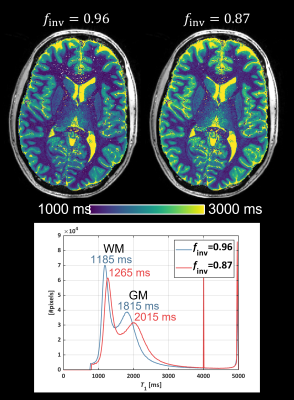 |
Mapping the inversion efficiency (finv): Comparison between adiabatic pulses for use in MP2RAGE-based T1-mapping
Hampus Olsson1 and Gunther Helms1
1Medical Radiation Physics, Lund University, Lund, Sweden
Accuracy in MP2RAGE-based T1-mapping relies on knowledge of the imperfect inversion efficiency (finv) of the inversion pulse. Here we map the inversion efficiency of different adiabatic inversion pulses in human brain at 7T. We identified a mean finv=0.87, which is substantially lower than the finv=0.96 typically assumed in MP2RAGE. We further observed a lower finv in white matter (WM) than in the rest of the cerebrum. Subsequent T1 estimates differed by approximately 80 ms in WM and 200 ms in gray matter (GM) when assuming either finv=0.87 or finv=0.96.
|
|
| 17:01 | 0373.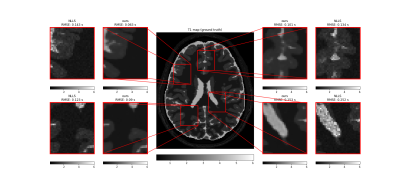 |
Denoising and Uncertainty Estimation in parameter mapping With Bayesian Deep Image Prior
Max Hellström1 and Anders Garpebring1
1Radiation Sciences, Umeå University, Umeå, Sweden
Tissue parameter estimation often gives noisy parameter maps due to noise in the signal data. In this work, we improve parameter mapping by incorporating noise reduction and uncertainty estimation with Bayesian Deep Image Prior. We implement task-specific loss functions for different applications. We test our method by estimating T1, T2, and ADC with synthetic- and in-vivo MRI data. Our method results in denoised tissue parameter maps, with associated error estimates. Our method is easy to implement, does not require any training data, and is easy to customize for different applications in parameter mapping.
|
|
| 17:03 | 0374.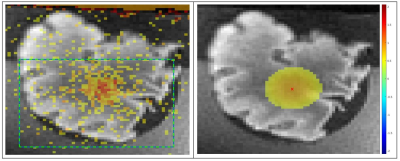 |
MR-Thermometry based targeting for histotripsy treatments in ex-vivo tissues
Dinank Gupta1, Dave Choi1, Steven Allen2, Tim Hall1, Zhen Xu1, and Douglas Noll1
1University of Michigan, Ann Arbor, MI, United States, 2Brigham Young University, Provo, UT, United States
The accuracy of using MR-Thermometry for pre-treatment targeting for histotripsy was evaluated on ex-vivo tissues. Eight brain samples were treated with an ultrasound array capable of performing both heating and histotripsy treatments. Heating was performed at array focus while MR-Thermometry images were acquired. Subsequently, histotripsy lesions were also made at the focus and post-treatment images were acquired. The location of MR-Thermometry estimated focal-zone was compared with the observed histotripsy lesion location to assess the accuracy of MR-Thermometry targeting. The mean error in estimated focus was about 1mm in all axes, suggesting MR-Thermometry would be suitable for pre-treatment targeting for histotripsy.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.