Combined Educational & Scientific Session
Artificial Intelligence/Machine Learning New Technology & Clinical Translation
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 08:15 | AI/ML Clinical Translation & Impact Greg Zaharchuk | |
| 08:35 | AI/ML Emerging Technologies & Innovative Methods
Mehmet Akçakaya
Keywords: Image acquisition: Machine learning, Image acquisition: Image processing, Image acquisition: Reconstruction Artificial intelligence/machine learning (AI/ML) based techniques have gathered interest as a possible means to improve MRI processing pipelines, with applications ranging from image reconstruction from raw data to extraction of quantitative biomarkers from imaging data in post-processing. Our purpose is to review existing and emerging AI/ML methods for various MRI processing applications. |
|
| 08:55 | 1164.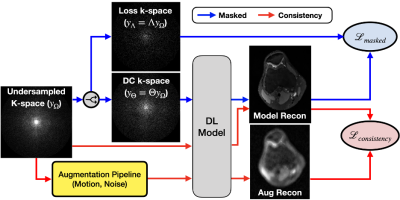 |
VORTEX-SS: Encoding Physics-Driven Data Priors for Robust Self-Supervised MRI Reconstruction
Arjun Desai1,2, Beliz Gunel1, Batu Ozturkler1, Brian A Hargreaves1,2, Garry E Gold2, Shreyas Vasanawala2, John Pauly1, Christopher Ré3, and Akshay S Chaudhari2,4
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3Computer Science, Stanford University, Stanford, CA, United States, 4Biomedical Data Science, Stanford University, Stanford, CA, United States Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Artifacts Deep learning (DL) has demonstrated promise for fast, high quality accelerated MRI reconstruction. However, current supervised methods require access to fully-sampled training data, and self-supervised methods are sensitive to out-of-distribution data (e.g. low-SNR, anatomy shifts, motion artifacts). In this work, we propose a self-supervised, consistency-based method for robust accelerated MRI reconstruction using physics-driven data priors (termed VORTEX-SS). We demonstrate that without any fully-sampled training data, VORTEX-SS 1) achieves high performance on in-distribution, artifact-free scans, 2) improves reconstructions for scans with physics-driven perturbations (e.g. noise, motion artifacts), and 3) generalizes to distribution shifts not modeled during training. |
| 09:03 | 1165.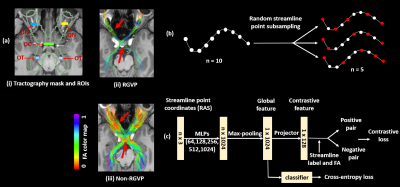 |
A novel deep learning method for automated identification of the retinogeniculate pathway using dMRI tractography
Sipei Li1,2, Jianzhong He2,3, Tengfei Xue2,4, Guoqiang Xie2,5, Shun Yao2,6, Yuqian Chen2,4, Erickson F. Torio2, Yuanjing Feng3, Dhiego CA Bastos2, Yogesh Rathi2, Nikos Makris2,7, Ron Kikinis2, Wenya Linda Bi2, Alexandra J Golby2, Lauren J O’Donnell2, and Fan Zhang2
1University of Electronic Science and Technology of China, Chengdu, China, 2Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States, 3Zhejiang University of Technology, Hangzhou, China, 4University of Sydney, Sydney, Australia, 5Nuclear Industry 215 Hospital of Shaanxi Province, Xianyang, China, 6The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China, 7Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States Keywords: Nerves, Brain We present a novel deep learning framework, DeepRGVP, for the retinogeniculate pathway (RGVP) identification from dMRI tractography data. We propose a novel microstructure-supervised contrastive learning method (MicroSCL) that leverages both streamline labels and tissue microstructure (fractional anisotropy) for RGVP and non-RGVP. We propose a simple and effective streamline-level data augmentation method (StreamDA) to address highly imbalanced training data. We perform comparisons with three state-of-the-art methods on an RGVP dataset. Experimental results show that DeepRGVP has superior RGVP identification performance. |
09:11 |
1166.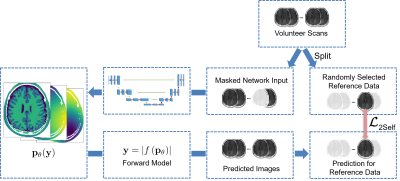 |
Semi-Supervised Learning for Spatially Regularized Quantitative MRI Reconstruction - Application to Simultaneous T1, B0, B1 Mapping
Felix Frederik Zimmermann1, Andreas Kofler1, Christoph Kolbitsch1, and Patrick Schuenke1
1Physikalisch-Technische Bundesanstalt (PTB), Berlin and Braunschweig, Germany Keywords: Quantitative Imaging, Machine Learning/Artificial Intelligence Typically, in quantitative MRI, an inverse problem of finding parameter maps from magnitude images has to be solved. Neural networks can be applied to replace non-linear regression models and implicitly learn a suitable spatial regularization. However, labeled training data is often limited. Thus, we propose a combination of training on synthetic data and on unlabeled in-vivo data utilizing pseudo-labels and a Noise2Self-inspired technique. We present a convolutional neural network trained to predict T1, B0, and B1 maps and their estimated aleatoric uncertainties from a single WASABITI scan. |
09:19 |
1167.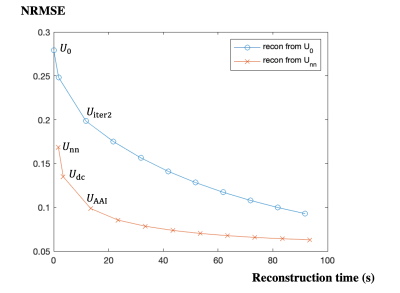 |
AI-Assisted Iterative Reconstruction for CMR Multitasking
Zihao Chen1,2, Hsu-Lei Lee1, Yibin Xie1, Debiao Li1,2, and Anthony Christodoulou1,2
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence CMR Multitasking is a promising approach for quantitative imaging without breath-holds or ECG monitoring but standard iterative reconstruction is too long for clinical use. Supervised artificial intelligence (AI) can accelerate reconstruction but lacks generalizability and transparency, and T1 mapping precision has not been sufficient. Here we propose an AI-Assisted Iterative (AAI) reconstruction which takes an AI reconstruction output as a “warm start” to a well-characterized iterative reconstruction algorithm with only 2 iterations. The proposed method produces better image fidelity and more precise T1 maps than other accelerated reconstruction methods, in less than 15 seconds (16x faster than conventional iterative reconstruction). |
| 09:27 | 1168.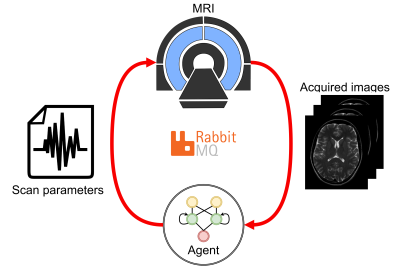 |
Feasibility of automatic patient-specific sequence optimization with deep reinforcement learning
Maarten Terpstra1,2, Sjors Verschuren1,2, Tom Bruijnen1,2, Matteo Maspero1,2, and Cornelis van den Berg1,2
1Department of Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 2Computational Imaging Group for MR Diagnostics & Therapy, University Medical Center Utrecht, Utrecht, Netherlands Keywords: Pulse Sequence Design, Machine Learning/Artificial Intelligence, Autonomous sequence optimization For MRI-guided interventions, tumor contrast and visibility are crucial. However, the tumor tissue parameters can significantly vary among subjects, with a range of T1, T2, and proton density values that may cause sub-optimal image quality when scanning with population-optimized protocols. Patient-specific sequence optimization could significantly increase image quality, but manual parameter optimization is infeasible due to the high number of parameters. Here, we propose to perform automatic patient-specific sequence optimization by applying deep reinforcement learning and reaching near-optimal SNR and CNR with minimal additional acquisitions. |
| 09:35 | 1169.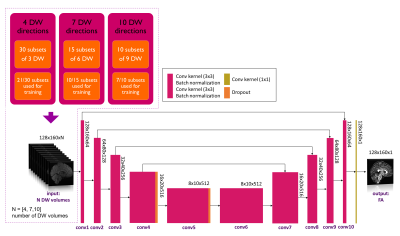 |
Attention please: deep learning can reproduce fractional anisotropy microstructure maps with reduced input data but loses clinical sensitivity
Marta Gaviraghi1, Antonio Ricciardi2, Fulvia Palesi1, Wallace Brownlee2, Paolo Vitali3,4, Ferran Prados2,5,6, Baris Kanber2,5, and Claudia A. M. Gandini Wheeler-Kingshott1,2,7
1Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 2NMR Research Unit, Department of Neuroinflammation, Queen Square Multiple Sclerosis Centre, UCL Queen Square Institute of Neurology, University College London (UCL), London, United Kingdom, 3Department of Radiology, IRCCS Policlinico San Donato, Milan, Italy, 4Department of Biomedical Sciences for Health, Università degli Studi di Milano, Milan, Italy, 5Department of Medical Physics and Biomedical Engineering, Centre for Medical Image Computing (CMIC), University College London, London, United Kingdom, 6Universitat Oberta de Catalunya, E-Health Center, Barcelona, Spain, 7Brain Connectivity Centre, IRCCS Mondino Foundation, Pavia, Italy Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, fractional anisotropy Quantitative maps obtained with diffusion weighted (DW) imaging such as fractional anisotropy (FA) are useful in pathologies. Often, to speed up acquisition time, the number of DW volumes acquired is reduced. We investigated the performance and clinical sensitivity of deep learning (DL) networks to calculate FA starting from different numbers of DW volumes. Using 4 or 7 volumes, clinical sensitivity was affected because no consistent differences between groups were found, contrary to our “one-minute FA” that uses 10 DW volumes. When developing DL for reduced acquisition data, the ability to generalize and biomarker sensitivity must be assessed. |
| 09:43 | 1170.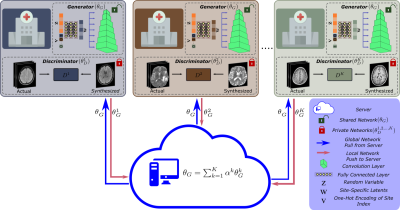 |
Federated MRI Reconstruction with Deep Generative Models
Gokberk Elmas1,2, Salman Ul Hassan Dar1,2, Yilmaz Korkmaz1,2, Muzaffer Ozbey1,2, and Tolga Cukur1,2,3
1Department Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence Generalization performance in learning-based MRI reconstruction relies on comprehensive model training on large, diverse datasets collected at multiple institutions. Yet, centralized training after cross-site transfer of imaging data introduces patient privacy risks. Federated learning (FL) is a promising framework that enables collaborative training without explicit data sharing across sites. Here, we introduce a novel FL method for MRI reconstruction based on a multi-site deep generative model. To improve performance and reliability against data heterogeneity across sites, the proposed method decentrally trains a generative image prior decoupled from the imaging operator, and adapts it to minimize data-consistency loss during inference. |
| 09:51 | 1171.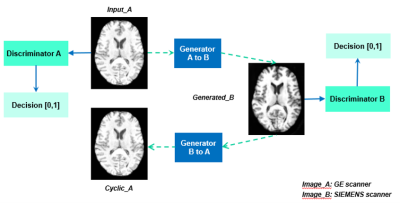 |
Unsupervised learning for MRI cross-scanner harmonization
Grace Wen1, Vickie Shim1,2, Miao Qiao3, Justin fernandez1,2, Samantha Holdsworth2,4, and Alan Wang1,4 1Auckland Bioengineering Institue, university of Auckland, Auckland, New Zealand, 2Mātai Medical Research Institute, Gisborne, New Zealand, 3Department of Computer Science, Faculty of Science, University of Auckland, Auckland, New Zealand, 4Department of Anatomy and Medical Imaging, Faculty of Medical and Health Sciences, University of Auckland, Auckland, New Zealand Keywords: Multi-Contrast, Machine Learning/Artificial Intelligence, harmonization, normalization, reconstruction Harmonization is necessary for large-scale multi-site neuroimaging studies to reduce the variations due to factors such as image acquisition, imaging devices, and acquisition protocols. This so-called scanner effect significantly impacts multivariate analysis and the development of computational predictive models using MRI. Our approach utilized an unsupervised learning based model to build a mapping between MR data acquired from two different scanners. Results illustrate the potential of unsupervised deep learning algorithms to harmonize MRI data, as well as to improve downstream tasks by applying the harmonization. |
| 09:59 | 1172.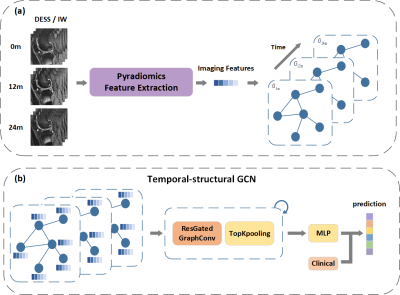 |
Temporal-Structural Graph Convolutional Network for Knee Osteoarthritis Progression Prediction Using MRI from the Osteoarthritis Initiative
Jiaping Hu1, Zidong Zhou2,3, Junyi Peng2,3, Lijie Zhong1, Kexin Jiang1, Zhongping Zhang4, Lijun Lu2,3,5, and Xiaodong Zhang1 1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, GuangZhou, China, 2School of Biomedical Engineering and Guangdong Provincial Key Laboratory of Medical Image Processing, Southern Medical University, GuangZhou, China, 3Guangdong Province Engineering Laboratory for Medical Imaging and Diagnostic Technology, Southern Medical University, Guangzhou, China, 4Philips Healthcare, GuangZhou, China, 5Pazhou Lab, Guangzhou, China Keywords: Osteoarthritis, Machine Learning/Artificial Intelligence Identifying patients with knee osteoarthritis (OA) whom the disease will progress is critical in clinical practice. Currently, the time-series information and interactions between the structures and sub-regions of the whole knee are underused for predicting. Therefore, we propose a temporal-structural graph convolutional network (TSGCN) using time-series data of 194 cases and 406 OA comparators. Each sub-region was regarded as a vertex and represented by the extracted radiomics features, the edges between vertexs were established by the clinical prior knowledge. The multiple-modality TSGCN (integrating information of MRIs, clinical and image-based semi-quantitative score) performed best comparing to the radiomics and CNN model. |
10:07 |
1173.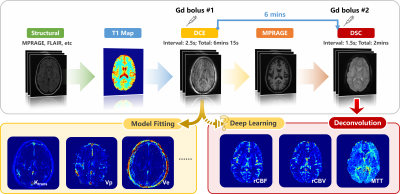 |
DSC-derived perfusion map generation from DCE MRI using deep learning
Haoyang Pei1,2, Yixuan Lyu2,3, Sebastian Lambrecht4,5,6, Doris Lin5, Li Feng1, Fang Liu7, Paul Nyquist8, Peter van Zijl5,9, Linda Knutsson5,9,10, and Xiang Xu1,5
1Biomedical Engineering and Imaging Institute and Department of Radiology, Icahn School of Medicine at Mount Sinai, New York City, NY, United States, 2Department of Electrical and Computer Engineering, NYU Tandon School of Engineering, New York City, NY, United States, 3Image Processing Center, School of Astronautics, Beihang University, Beijing, China, 4Department of Neurology, Technical University of Munich, Munich, Germany, 5Department of Radiology, Johns Hopkins University, Baltimore, MD, United States, 6Institute of Neuroradiology, Ludwig-Maximilians-Universität, Munich, Germany, 7Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States, 8Department of Neurology, Johns Hopkins University, Baltimore, MD, United States, 9F.M Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 10Department of Medical Radiation Physics, Lund University, Lund, Sweden Keywords: Machine Learning/Artificial Intelligence, Perfusion This study built a deep-learning-based method to directly extract DSC MRI perfusion and perfusion related parameters from DCE MRI. A conditional generative adversarial network was modified to solve the pixel-to-pixel perfusion map generation problem. We demonstrate that in both healthy and brain tumor patients, highly realistic perfusion and perfusion related parameter maps can be synthesized from the DCE MRI using this deep-learning method. In healthy controls, the synthesized parameters had distribution similar to the ground truth DSC MRI values. In tumor regions, the synthesized parameters correlated linearly with the ground truth values. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.