Digital Poster
Quality, Reproducibility & Harmony
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
3233.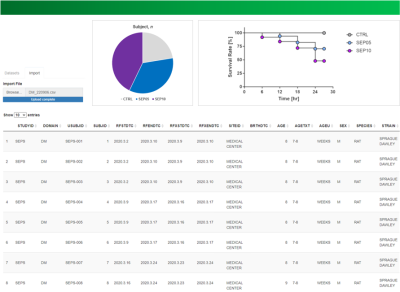 |
1 |
Conversion of legacy data of nonclinical trials using MRI into
CDISC SEND: Toward Standardization of an Animal MR Imaging Data
Do-Wan Lee1,
Young Chul Cho2,
Mi-hyun Kim3,4,
Yeon Ji Chae5,
Su Jung Ham3,
Yousun Ko1,
Seongwon Na2,
Youngbin Shin2,
Nari Kim6,
Dong‐Cheol Woo5,7,
and Kyung Won Kim2
1Department of Radiology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 2Biomedical Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of, 3Trialinformatics Inc., Seoul, Korea, Republic of, 4Department of Radiation Science & Technology, Jeonbuk National University, Jeonju, Korea, Republic of, 5Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 6Department of Medical Science, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 7Convergence Medicine Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of Keywords: Visualization, Visualization In nonclinical animal research for new drug development, the US FDA mandates to manage data in accordance with clinical data interchange standards consortium (CDISC) standard for exchange of nonclinical data (SEND). Older nonclinical trial data, i.e., legacy nonclinical trial data, contain highly useful information but those data were created in various data formats by different researchers. The present study specifically describes how to implement the CDISC SEND data standards for the legacy nonclinical trial data with MRI. Standardization of animal MRI data with CDISC SEND enables us to utilize legacy data for next-generation drug development and new knowledge creation. |
|
3234.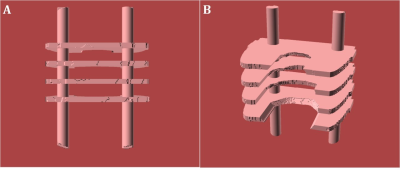 |
2 |
IMPROVED EX-VIVO IMAGING USING BRAIN & CONTAINER-SPECIFIC
3D-PRINTED HOLDERS
Sethu K. Boopathy Jegathambal1,
Ilana Ruth Leppert2,
David A. Rudko2,
and Amir Shmuel2
1Department of Biomedical Engineering, McGill University, Montreal, QC, Canada, 2McConnell Brain Imaging Center, Montreal Neurological Institute and Hospital, Montreal, QC, Canada Keywords: Data Acquisition, Ex-Vivo Applications, Brain Ex-Vivo MRI Depending on the medium, it might be difficult to maintain the brain stable during ex-vivo imaging. In addition, the SNR might not be homogeneous. We propose using brain and container-specific 3D-printed models for holding the brain. The method requires preliminary imaging of the container and the brain. Then, a 3D-printed model is created, with profiles that match the surface of the brain on one side and the inner surface of the container on the other. The holder is designed to maintain the brain stable in space, where the SNR is high and homogeneous, while allowing contact with the immersion fluid. |
|
3235.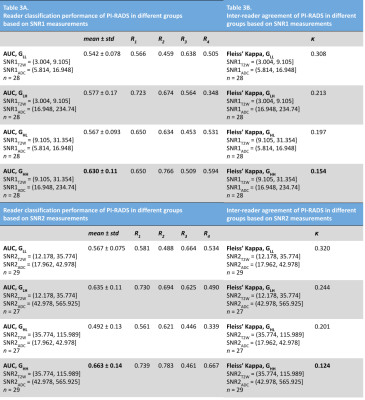 |
3 |
Association of MR image quality measures with diagnostic
accuracy and inter-reader agreement of PI-RADS for detection of
prostate cancer
Pranav Sompalle1,
Ansh Roge1,
Michael Sobota1,
Amogh Hiremath2,
Sree Harsha Tirumani3,
Leonardo Kayat Bittencourt3,
Ryan Ward4,
Justin Ream4,
Andrei Purysko4,
Anant Madabhushi5,
Satish Viswanath1,
and Rakesh Shiradkar5
1Case Western Reserve University, Cleveland, OH, United States, 2Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 3University Hospitals Cleveland Medical Center, Cleveland, OH, United States, 4Cleveland Clinic Foundation, Cleveland, OH, United States, 5Emory University, Atlanta, GA, United States Keywords: Data Processing, Data Processing, Image processing Radiologists’ ability to detect clinically-significant prostate cancer on MRI is affected by image quality. While subjective guidelines are provided by the prostate imaging reporting and data system (PI-RADS) and other recent methods, there is a need to develop objective and quantitative metrics of MRI quality. In this multi-reader study, we derive MRI quality metrics using an image processing-based open-source software tool and evaluate their association with radiologist-assigned PI-RADS scores to detect clinically-significant prostate cancer (csPCa). We observe that higher quality MRI is associated with improved detection of csPCa on MRI. |
|
3236.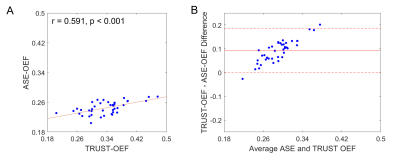 |
4 |
Dual-site and dual-vendor comparison of cerebral oxygen
extraction fraction by ASE and TRUST MRI in identical
participants
Chunwei Ying1,
Spencer Waddle2,
Nkemdilim Igwe3,
Niral J. Patel4,
Alexander K. Song2,
Charu Balamurugan4,
Lori C. Jordan4,
Dengrong Jiang5,
Hanzhang Lu5,
Andria L. Ford3,
Manus J. Donahue2,
and Hongyu An1,3
1Mallinckrodt Institute of Radiology, Washington University School of Medicine, St Louis, MO, United States, 2Department of Neurology, Vanderbilt University Medical Center, Nashville, TN, United States, 3Department of Neurology, Washington University School of Medicine, St Louis, MO, United States, 4Department of Pediatrics, Vanderbilt University Medical Center, Nashville, TN, United States, 5Department of Radiology, Johns Hopkins University School of Medicine, Baltimore, MD, United States Keywords: Data Analysis, Brain, repeatability; reproducibility We evaluated dual-site and dual-vendor test-retest repeatability and reproducibility of cerebral oxygen extraction fraction (OEF) measured with asymmetric spin echo (ASE) and T2-Relaxation-Under-Spin-Tagging (TRUST) MRI. The intra-site intra-class correlation coefficient (ICC) of global ASE-OEF were 0.952 and 0.919 for each site. The intra-site ICC of TRUST-OEF based on the bovine and the HbA model were 0.810 and 0.792 at site 1, and 0.928 and 0.924 at site 2. The inter-site inter-vendor correlation was 0.849 for ASE-OEF and 0.814 for TRUST-OEF based on the bovine model. ASE-OEF was significantly associated with TRUST-OEF (p<0.001). |
|
3237.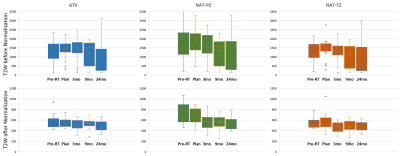 |
5 |
A Novel Deep Learning Tissue Normalization Method for
Longitudinal Analysis of T2-Weighted MRI following Prostate
Cancer Radiation Treatment
Ahmad Algohary1,
Evangelia I. Zacharaki1,
Mohammad Alhusseini1,
Adrian Breto1,
Veronica Wallaengen1,
Isaac Xu1,
Sandra Gaston1,
Patricia Castillo1,
Sanoj Punnen1,
Benjamin Spieler1,
Matthew Abramowitz1,
Alan Dal Pra1,
Oleksandr Kryvenko1,
Alan Pollack1,
and Radka Stoyanova1
1The University of Miami, Miami, FL, United States Keywords: Machine Learning/Artificial Intelligence, Prostate In this work, we introduce a novel automated triple-reference intensity normalization method for T2W images with the aim of obtaining consistent longitudinal measurements leading to improved quantitative assessment of radiation treatment outcome for prostate cancer patients. |
|
3238.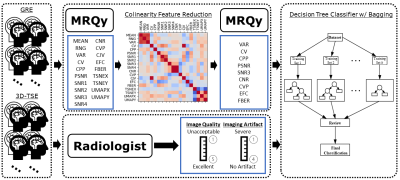 |
6 |
The Use of Quantitative Metrics and Machine Learning to Predict
Radiologist Interpretations of Image Quality and Artifacts
Lucas McCullum1,
John Wood2,
Maria Gule-Monroe3,
Ho-Ling Anthony Liu4,
Melissa Chen3,
Komal Shah3,
Noah Nathan Chasen3,
Vinodh Kumar3,
Ping Hou4,
Jason Stafford4,
Caroline Chung5,
Moiz Ahmad4,
Christopher Walker4,
and Joshua Yung4
1Medical Physics, MD Anderson Cancer Center, Houston, TX, United States, 2Enterprise Development and Integration, MD Anderson Cancer Center, Houston, TX, United States, 3Neuroradiology, MD Anderson Cancer Center, Houston, TX, United States, 4Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 5Data Governance & Provenance, MD Anderson Cancer Center, Houston, TX, United States Keywords: Machine Learning/Artificial Intelligence, Data Analysis, Image Quality A dataset of 3D-GRE and 3D-TSE brain 3T post contrast T1-weighted images as part of a quality improvement project were collected and shown to five neuro-radiologists who evaluated each sequence for image quality and artifacts. The same scans were processed using the MRQy tool for objective, quantitative image quality metrics. Using the combined radiologist and quantitative metrics dataset, a decision tree classifier with a bagging ensemble approach was trained to predict radiologist assessment using the quantitative metrics. The resulting AUCs for each classification task were above 0.7 for all combinations of sufficiently represented classes and qualitative image metrics. |
|
3239.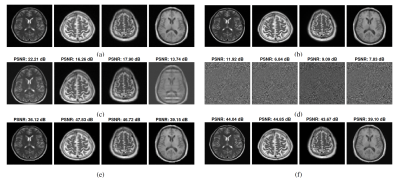 |
7 |
Guided field Map Estimation and Image Correction for MRI with
LargeMagnetic Field Inhomogeneity
Navid Reyhanian1,
Michael Mullen2,
Taylor Froelich2,
Michael Garwood2,
and Jarvis Haupt1
1Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States Keywords: Data Analysis, Magnets (B0), optimization; static field inhomogeneity; artifact correction Many methods deployed for Magnetic Resonance Imaging (MRI) suffer from degraded image quality when the static magnetic field ($$$B_0$$$) is nonuniform, as $$$B_0$$$ inhomogeneity causes different types of image artifacts and distortions. Image correction methods require a prior estimate of the field map. In this work, we study the problem of static $$$B_0$$$ inhomogeneity estimation from distorted and undistorted image pairs, despite undistorted images being noisy. The estimated field map is later utilized to correct distorted images. The efficacy of the proposed approach is demonstrated for different encoding methods in the presence of large magnetic field inhomogeneity via extensive simulations. |
|
3240.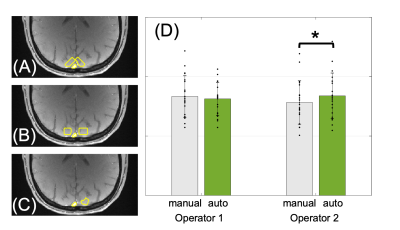 |
8 |
Semi-automated analysis to reduce operator variability in oxygen
extraction fraction using susceptometry-based oximetry
Bo-Jhen Su1,
Pei-Hsin Wu1,
Rajiv S Deshpande2,
and Felix W Wehrli2
1Department of Electrical Engineering, National Sun Yat-sen University, Kaohsiung, Taiwan, 2Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States Keywords: Data Analysis, Oxygenation SvO2 and OEF were estimated via susceptometry-based oximetry (SBO). The impact of user-defined ROI was investigated in this study. The results show that (1) the consistency of SvO2 estimation depends on operators’ comprehension of MR-oximetry, (2) SvO2 might be under-estimated with manual ROI selection, and (3) the operator-induced bias can be eliminated by the semi-automatic analysis. The preliminary data indicate the feasibility of the constructed semi-automatic analysis for consistent estimation of SvO2 among operators and its potential for bulk analysis and clinical use. |
|
3241.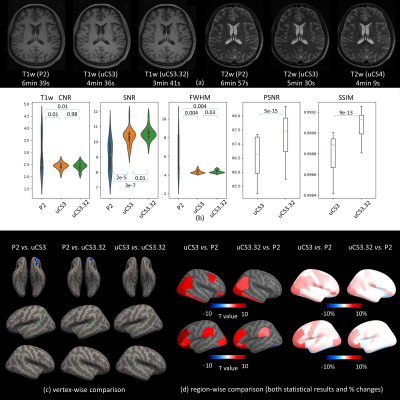 |
9 |
A Comprehensive Quality Assessment on Fast MRI Acquisition with
united Compressed Sensing Techniques
Zhuoyang Gu1,
Lianghu Guo1,
Qing Yang1,
Xinyi Cai1,
Tianli Tao1,
Sifan He1,
Qian Wang1,
He Qiang2,
Dinggang Shen1,
and Han Zhang1
1School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 2United Imaging Healthcare Co., Ltd., Shanghai, China Keywords: Parallel Imaging, Data Acquisition, Quality Assessment MRI acquisition from certain populations is difficult. For instance, infant MRI without sedation during wakefulness has a high failure rate due to head motion. Thus, a shorter scan time is required. Increasing studies focused on fast MRI acquisition but few studies examined their qualities. We argue that only calculating common image quality metrics (e.g., SNR) may not meet the satisfaction of brain MRI analysis with fast acquisition techniques. A whole-process quality assessment method is thus proposed comparing different fast acquisition techniques with practical guidelines. In future, comprehensive quality assessment on fast MRI acquisitions can become a study routine. |
|
3242.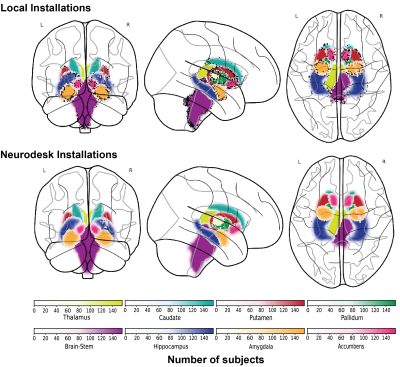 |
10 |
Investigating the computational reproducibility of Neurodesk
Thuy Thanh Dao1,
Angela Renton1,2,
Aswin Narayanan3,
Markus Barth1,
and Steffen Bollmann1
1School of Information Technology and Electrical Engineering, The University of Queensland, Brisbane, Australia, 2Queensland Brain Institute, The University of Queensland, Brisbane, Australia, 3Australian National Imaging Facility, The University of Queensland, Brisbane, Australia Keywords: Software Tools, Data Processing Glatard et al [1] found that the operating system version affected the computational results of neuroimaging analysis pipelines. Their work demonstrated that this reproducibility issue is caused by the accumulated floating-point differences across operating systems. Neurodesk (www.neurodesk.org) approaches this problem by packaging every pipeline into a software container and thereby can control the underlying dependencies for each application regardless of the operating system. In this work, a brain tissue segmentation pipeline from the FMRIB Software Library (FSL) was compared with a containerised version from Neurodesk. The comparison revealed that Neurodesk provides computational reproducibility compared with local installations. |
|
3243.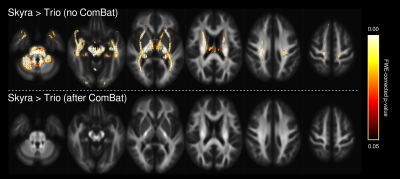 |
11 |
ComBat scanner harmonization for fixel-based analysis
Remika Mito1,2,
Heath Pardoe1,
Robert Smith1,
Jacques-Donald Tournier3,4,
David Vaughan1,2,5,
Mangor Pedersen6,
and Graeme Jackson1,2,5
1Florey Institute of Neuroscience and Mental Health, Melbourne, Australia, 2Florey Department of Neuroscience and Mental Health, University of Melbourne, Melbourne, Australia, 3Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 4Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 5Department of Neurology, Austin Health, Melbourne, Australia, 6Auckland University of Technology, Auckland, New Zealand Keywords: Data Analysis, Diffusion/other diffusion imaging techniques, Data harmonization Diffusion MRI data are known to be sensitive to scanner and site effects, for which harmonization methods have been developed. However, harmonization methods have not yet been applied to many advanced diffusion imaging metrics. In this work, we apply the ComBat harmonization approach to fixel-based analysis. ComBat successfully minimizes scanner-related differences in fibre density and cross-section, and performs similarly to the inclusion of scanner as a nuisance regressor during whole-brain fixel-based analysis. Importantly, ComBat can now readily be used within the fixel-based framework, which will enable large multi-centre studies to implement this approach in the future. |
|
3244.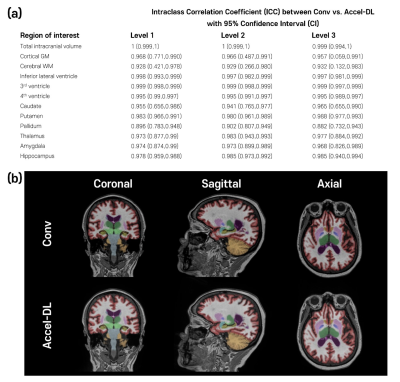 |
12 |
Reliability of brain volume measures of accelerated 3D
T1-weighted images with deep learning-based reconstruction
Woojin Jung1,
Seongjae Mun1,
Jingyu Ko1,
and Koung Mi Kang2
1AIRS Medical, Seoul, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Brain We validated the brain volumetric results with 3D T1 weighted images with accelerated scans reconstructed by FDA-cleared deep learning-based software (SwiftMR, AIRS Medical). Acceleration scans with three different acceleration levels were simulated using k-space undersampling, and the image quality and brain volume measures were evaluated. In addition, we acquired conventional and accelerated scans from each participant to evaluate the reliability between conventional and accelerated scans reconstructed by SwiftMR and inter-method reliability between different brain segmentation software. As a result, brain volume measures with accelerated scans with deep learning-based reconstruction were in good agreement with those of the corresponding conventional scan. |
|
3245.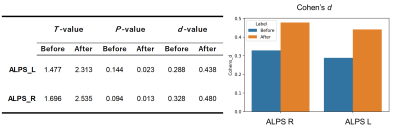 |
13 |
Harmonization of the multisite ALPS-index based on DTI-ALPS
using the combined association test
Yuya Saito1,
Koji Kamagata1,
Toshiaki Taoka2,
Christina Andica1,
Wataru Uchida1,
Kaito Takabayashi1,
Mana Owaki1,3,
Seina Yoshida1,3,
Keigo Yamazaki1,3,
Shohei Fujita1,4,
Akifumi Hagiwara1,
Junko Kikuta1,
Toshiaki Akashi1,
Akihiko Wada1,
Keigo Shimoji1,
Masaaki Hori5,
Shinji Naganawa6,
and Shigeki Aoki1
1Department of Radiology, Juntendo University Graduate School of Medicine, Tokyo, Japan, 2Department of Innovative Biomedical Visualization (iBMV), Nagoya University Graduate School of Medicine, Aichi, Japan, 3Department of Radiological Sciences, Graduate School of Human Health Sciences, Tokyo Metropolitan University, Tokyo, Japan, 4Department of Radiology, University of Tokyo, Tokyo, Japan, 5Department of Radiology, Toho University Omori Medical Center, Tokyo, Japan, 6Department of Radiology, Nagoya University Graduate School of Medicine, Aichi, Japan Keywords: Data Processing, Data Processing, Diffusion MRI, ALPS-index, DTI-ALPS, Harmonization, multisite study Diffusion tensor image analysis along the perivascular space (DTI-ALPS) is a promising noninvasive method for indirectly evaluating the glymphatic system. However, the ALPS-index calculated from diffusion MRI data collected at multiple sites should be harmonized to avoid site-related effects. We applied the combined association test (ComBat), which uses regression of covariates with empirical Bayes, for harmonizing the ALPS-index. ComBat mitigated site-related effects, increased statistical power to differentiate Alzheimer’s disease and cognitive normal, and improved the correlation between the ALPS-index and cognitive function. Thus, ComBat harmonization can be applied to evaluate the glymphatic system using the ALPS-index in large multisite studies. |
|
3246.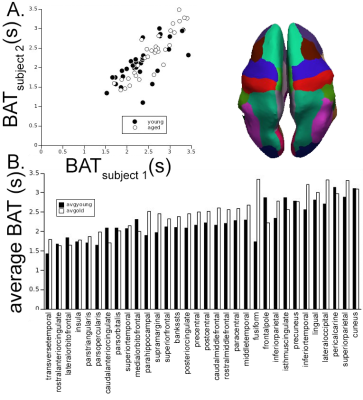 |
14 |
Reliable and repeatable fully automated bolus arrival times and
arterial inputs using 2 Hz whole brain multiband DSC-MRI at 7T
Daniel L Schwartz1,
Xin Li2,
Fred Lesage3,
Andreas Linninger4,
Mohammad Jamshidi4,
Becky Stone5,
and William Rooney5
1Advanced Imaging Research Center/Neurology, OHSU, PORTLAND, OR, United States, 2Advanced Imaging Research Center, OHSU, PORTLAND, OR, United States, 3Electrical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 4Bioengineering, UIC, Chicago, IL, United States, 5Advanced Imaging Research Center, OHSU, Portland, OR, United States Keywords: Data Processing, DSC & DCE Perfusion, Whole-brain bolus arrival times Dynamic susceptibility contrast imaging has used bolus arrival time (BAT) in tissue, relative to an arterial input function (AIF), as a nuisance variable when deriving perfusion metrics (e.g. blood flow). However, BAT reflects physiological changes in both fluid dynamics of feeding vasculature and capillary inflow. This work presents a reproducible approach for reliable and repeatable quantification of an AIF and whole brain BAT, which itself may be germane to chronic and acute cerebrovascular disease (CVD), such as white matter hyperintensities, age-associated CVD, and stroke. |
|
3247.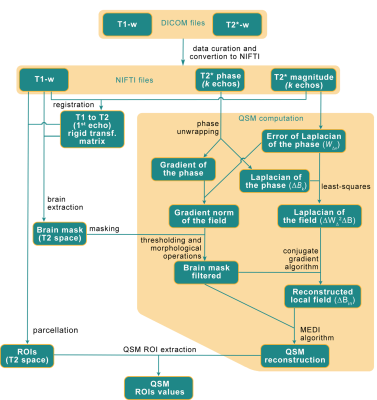 |
15 |
Automated brain QSM computation pipeline deployed in the
European Open Science Cloud
Miguel Guevara1,
Davy Cam2,
Jacques Badagbon2,
Stephane Roche2,
Michel Bottlaender1,
Yann Cointepas1,
Jean-François Mangin1,
Ludovic de Rochefort2,
and Alexandre Vignaud1
1Neurospin, Paris, France, 2Ventio, Marseille, France Keywords: Data Processing, Quantitative Susceptibility mapping, Cloud-computing Introduction of a pipeline for computing the Quantitative Susceptibility Mapping (QSM) from non-optimal MRI data in a secure cloud-infrastructure. The pipeline allows the computation of clean and exploitable QSM, thanks to a phase pre-processing to reduce the presence of artifacts in the 7T MRI data. |
|
| 3248. | 16 |
dbdicom: an open-source python interface for reading and writing
DICOM databases
Steven Sourbron1,
Joao Almeida e Sousa1,
Alexander Daniel2,
Charlotte Buchanan2,
Ebony Gunwhy1,
Eve Lennie1,
Kevin Teh1,
Steve Shillitoe1,
David Morris3,
Andrew Priest4,
David Thomas5,
and Susan Francis2
1University of Sheffield, Sheffield, United Kingdom, 2University of Nottingham, Nottingham, United Kingdom, 3University of Edinburgh, Edinburgh, Scotland, 4University of Cambridge, Cambridge, United Kingdom, 5University College London, London, United Kingdom Keywords: Software Tools, Data Processing DICOM is the universally recognized standard for medical imaging, but reading and writing from DICOM databases remains a challenging task for most data scientists. The dbdicom package was developed to provide an intuitive programming interface for reading and writing data from entire DICOM databases, and replaces confusing DICOM-native concepts by language and notations that will be more familiar to data scientists working in python. The package is available under an open license but is in the early stages of development and currently rolled out in 3 multi-vendor, multi-center studies. |
|
3249.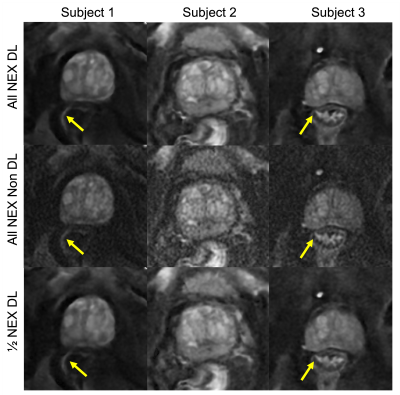 |
17 |
Improved Image Quality with Deep Learning-Based Image
Reconstruction for Multi-shot Diffusion-Weighted Imaging of the
Prostate
Patricia S. Lan1,
Xinzeng Wang2,
Alessandro Scotti3,
Praveen Jayapal4,
Pingni Wang1,
Arnaud Guidon5,
and Andreas M. Loening4
1GE Healthcare, Menlo Park, CA, United States, 2GE Healthcare, Houston, TX, United States, 3GE Healthcare, Columbus, OH, United States, 4Department of Radiology, Stanford University, Stanford, CA, United States, 5GE Healthcare, Boston, MA, United States Keywords: Diffusion/other diffusion imaging techniques, Diffusion/other diffusion imaging techniques Diffusion-weighted imaging (DWI) is a key component of identifying prostate tumors on MRI. Multi-shot DWI techniques (e.g. MUSE) have been shown to enable high resolution prostate DWI and, compared to single-shot DWI, reduce distortion artifacts due to rectal gas and hip implants at the expense of increased scan time. In this study, we evaluated a CNN-based deep learning (DL) image reconstruction method for MUSE. Our results indicate that for high b-value images the DL-based reconstruction improved perceived image quality even with half the original NEX, suggesting potential scan time reduction using DL. |
|
3250.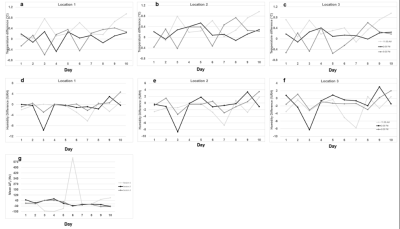 |
18 |
Repeatability of very low field MRI
Pavan Poojar1 and
Sairam Geethanath1
1Accessible Magnetic Resonance Laboratory, Biomedical Imaging and Engineering Institute, Department of Diagnostic, Molecular and Interventional Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, New York, NY, United States Keywords: Data Acquisition, Low-Field MRI Low-field MR scanners are low-cost, portable, and requires lesser space. In this study, we performed in vitro phantom repeatability study on a 48 mT scanner (Multiwave technologies) by scanning the phantom 3 sessions in a day (11 AM, 2 PM, and 5 PM) at 3 locations. During each scan, we measured the temperature, humidity, and F0 to see their effect on the image. We scanned 9 sequences in the study to monitor noise, F0 and B0. The difference in temperature and humidity was determined for each session.The line intensity profilesrevealed that VLF scanner produces consistent B0 maps and qualitative images. |
|
3251.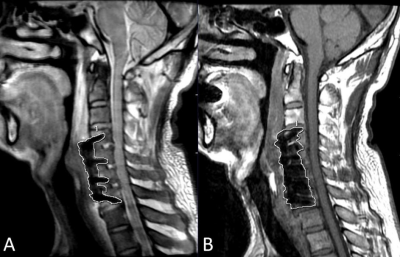 |
19 |
MAVRIC SL can reduce inter-rater variability of the measurements
for the cervical spine segment height in ACDF patients on 3T MRI
Renjie Yang1,
Changsheng Liu1,
Weiyin Vivian Liu2,
Liang Li1,
and Yunfei Zha1
1Renmin Hospital of Wuhan University, Wuhan, China, 2GE Healthcare, Beijing, China Keywords: Artifacts, Surgery Subsidence assessment is an important indicator for postoperative follow-ups for ACDF patients and normally performed on radiographs. It is still challenging to image ACDF patients using conventional MRI for the measure of segment height is influenced by the metal artifacts. In this study, MAVRIC SL showed better inter-rater agreement on measurements of segment height compared to FSE T1WI. 20 cases on MAVRIC SL and 6 cases on FSE T1W1 among 22 patients showed the measurements of segment height was less than the maximum allowed difference (2mm). This study demonstrated that MAVRIC SL has a strong potential in the subsidence assessment. |
|
3252.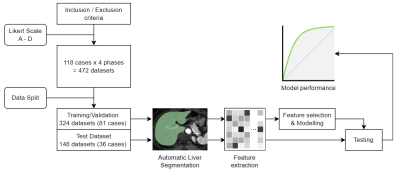 |
20 |
Automated Quality Assessment of Liver Magnetic Resonance Images
with Fully Automatic Segmentation and Radiomics Approach
Hai Nan Ren1,
Li Jun Qian1,
Xu Hua Gong1,
Yan Zhou1,
and Yang Song2
1Radiology, Renji Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China, 2MR Scientific Marketing, Siemens Healthineers Ltd., Shanghai, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Quality control This study investigated the feasibility and performance of quality assessment of hepatic magnetic resonance (MR) images using a deep-learning-based segmentation and radiomics approach. We used a pre-trained deep learning model to segment the liver on different contrast-enhanced MR phases and then extracted quantitative features to assess the image quality by a machine learning method. The results showed that the radiomics model had a high performance for image quality identification in both training and test sets. This suggests that it was feasible to automate the identification of image quality by using radiomics approaches. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.