Digital Poster
Image Reconstruction Methods I
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
4616.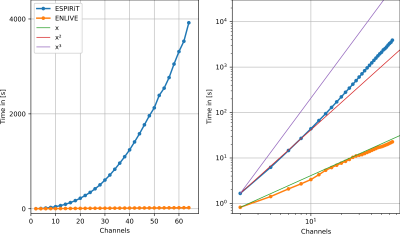 |
21 |
Non-Linear Reconstruction for Coil Sensitivity Calibration from
Cartesian and non-Cartesian Data
H. Christian M. Holme1 and
Martin Uecker1,2,3,4
1Institute of Biomedical Imaging, Graz University of Technology, Graz, Austria, 2Institute for Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 3Partner Site Göttingen, DZHK (German Centre for Cardiovascular Research), Göttingen, Germany, 4Cluster of Excellence “Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells” (MBExC), University of Göttingen, Göttingen, Germany Keywords: Parallel Imaging, Parallel Imaging We compare the current de-facto standard for coil sensitivity calculation, ESPIRiT, to a non-linear reconstruction method, ENLIVE. While ENLIVE normally produces both images and coil sensitivies, we focus here on using it for calibration of the coil sensitivities. We applied ESPIRiT and ENLIVE to low-resolution subsets of a 3D Cartesian and a non-Cartesian spiral acquisition, using the resoluting coil profiles in a linear parallel imaging reconstruction. We showed that ENLIVE is substantially faster for a high number of channels and provides improved sensitivities in the non-Cartesian example. |
|
4617.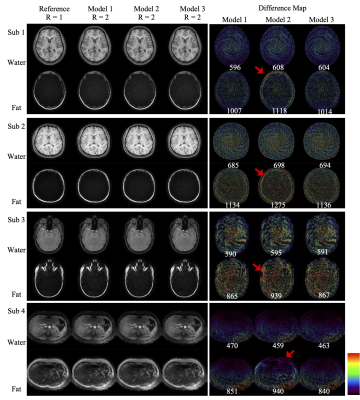 |
22 |
Fast and robust parallel imaging algorithm for Spiral MRI with
Fat-Water separation
Tzu Cheng Chao1,
Xi Peng1,
Dinghui Wang1,
and James G. Pipe1
1Department of Radiology, Mayo Clinic, Rochester, MN, United States Keywords: Parallel Imaging, Parallel Imaging Three different models to reconstruct fat and water spiral images with parallel acquisition are compared in terms of computational efficiency and reconstruction quality. The best reconstruction is obtained with a full model requiring the most computational time, while the model with the fastest computational time results in some degradation of image quality. A third, preferred model preserves most of the image quality of the first model, with nearly the same speedup as the latter model. |
|
4618.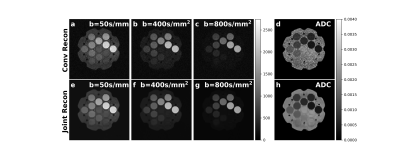 |
23 |
Joint K-b space reconstruction of under-sampled
diffusion-weighted MRI
Yan Dai1,
Jie Deng1,
and Xun Jia2
1University of Texas Southwestern Medical Center, Dallas, TX, United States, 2Johns Hopkins University, Baltimore, MD, United States Keywords: Image Reconstruction, Diffusion/other diffusion imaging techniques We developed a joint image reconstruction method in both k-space and b-space to reconstruct the under-sampled diffusion-weighted images acquired at different b-values and generate the corresponding apparent diffusion coefficient map simultaneously by solving an optimization problem. This method improved SNR in both diffusion weighted images and apparent diffusion coefficient map compared with the conventional method and allows 50% k-space data undersampling that has the potential of reducing image distortion and shortening acquisition time. |
|
4619. |
24 |
Time-Resolved MRI Kinesiology: Reconstructing force and velocity
during active loading
Max H.C. van Riel1,
Cornelis A.T. van den Berg1,
and Alessandro Sbrizzi1
1Department of Radiotherapy, Computational Imaging Group for MR Diagnostics and Therapy, UMC Utrecht, Utrecht, Netherlands Keywords: Signal Modeling, MSK, Time-Resolved, Velocity, Force, Motion Force and velocity are two important quantities for studying muscles, but retrieving these quantities using MRI is challenging. Spectro-Dynamic MRI allows the characterization of dynamical systems at a high spatial and temporal resolution directly from k-space data. Using an iterative reconstruction algorithm, we can reconstruct time-resolved MR images, time-resolved motion fields, mechanical parameters, and an activation force, at a temporal resolution of 11 ms. As a proof-of-principle, MR data was acquired with a motion phantom moving like an actively driven linear elastic system. All dynamic variables could be reconstructed accurately. This method could become useful for studying muscles under dynamic loads. |
|
4620.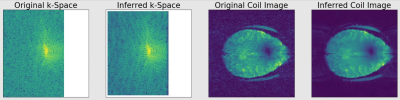 |
25 |
GRAPPA Interpolation with Unity Acceleration Enhances
Signal-to-Noise Ratio in Fully Sampled Acquisitions
Andrew S Nencka1,2
1Radiology, Medical College of Wisconsin, Milwaukee, WI, United States, 2Center for Imaging Research, Medical College of Wisconsin, Milwaukee, WI, United States Keywords: Image Reconstruction, Data Acquisition, De-noising Significant efforts are focused on maximizing SNR in high spatial and temporal resolution image acquisitions. We present a method of utilizing GRAPPA to de-noise k-space observations made with a multi-channel array. By replacing a fully sampled observed k-space with a k-space in which each observation is inferred from a weighted average of neighboring observations across all coils, an increase of SNR on the order of 5% is achieved. |
|
4621.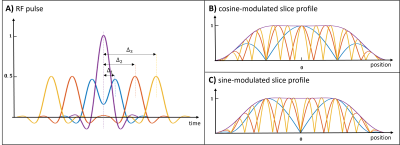 |
26 |
Spatially Fourier Excited Acquisition and Reconstruction (SFEAR)
for Improved Slice Acceleration
Negin Yaghmaie1,2,
Warda Syeda3,
Yasmin Blunck1,2,
Bahman Tahayori4,
Rebecca K. Glarin2,5,
Bradford A. Moffat2,6,
and Leigh A. Johnston1,2
1Department of Biomedical Engineering, The University of Melbourne, Melbourne, Australia, 2Melbourne Brain Centre Imaging Unit, The University of Melbourne, Melbourne, Australia, 3Melbourne Neuropsychiatry Centre, The University of Melbourne, Melbourne, Australia, 4The Florey Institute of Neuroscience and Mental Health, Melbourne, Australia, 5Department of Radiology, Royal Melbourne Hospital, Melbourne, Australia, 6Department of Medicine and Radiology, The University of Melbourne, Melbourne, Australia Keywords: New Trajectories & Spatial Encoding Methods, New Trajectories & Spatial Encoding Methods A new 3D Spatially Fourier Excited Acquisition and Reconstruction method (SFEAR) is introduced, in which double-RF pulses are used to excite sinusoidally-modulated slice profiles across a slab. Target spatial frequencies are achieved by varying the time shift between the two pulses, to construct the slice-phase-encode dimension of 3D k-space from Fourier excited acquisitions. SFEAR outperforms conventional GRAPPA in the slice-phase encode dimension, given its inherent ability to undersample sine or cosine components rather than whole planes of 3D k-space. Superior reconstruction of undersampled data and lower g-factor values are demonstrated in both 7T phantom and in vivo data. |
|
4622.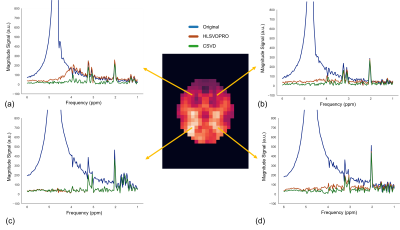 |
27 |
Water removal in MR spectroscopic imaging with Casorati Singular
Value Decomposition
Amirmohammad Shamaei1,
Jana Starcukova1,
Jedrek Burakiewicz 2,
and Zenon Starcuk 1
1Institute of Scientific Instruments of the Czech Academy of Sciences Research institute in Brno, Brno, Czech Republic, 2Tesla Dynamic Coils, Zaltbommel, Netherlands Keywords: Sparse & Low-Rank Models, Data Processing, Singular value decomposition, MR spectroscopic imaging, Water removal Removing residual water from the MRSI datasets using the SVD-based algorithms is computationally demanding. We present a novel algorithm to reduce the computing time required for water removal in MRSI data. Our proposed method exploits low-rank structures that exist in MRSI data. It arranges the MRSI data in the Casorati matrix form, applies singular value decomposition, and removes residual water from the most prominent left-singular vectors. We compared our proposed method with the HLSVDPRO method, and we achieved 20x acceleration while improving effectiveness. Our proposed method is publicly available as a pip-installable Python tool. |
|
4623.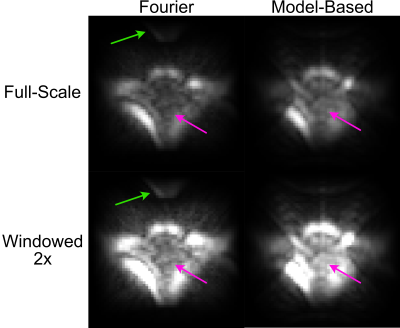 |
28 |
In Vivo xSPEN Imaging with a Model-Based Reconstruction for
Efficient Spatial Encoding in a Single-Sided Prostate MRI
Scanner
Muller De Matos Gomes1,
Meredith Sadinski1,
Alek Nacev1,
and William Allyn Grissom2
1Promaxo, Oakland, CA, United States, 2Biomedical Engineering, Vanderbilt University, Nashville, TN, United States Keywords: New Trajectories & Spatial Encoding Methods, Image Reconstruction, Low-Field MRI, Spatiotemporal Encoding Single-sided low-field MRI scanners can provide image guidance during interventions such as prostate biopsies without restricting surgical access, but efficient spatial encoding is a challenge. In this work we show how spatiotemporal encoding using the xSPEN method combined with a model-based image reconstruction enables swapping a conventionally phase encoded but large-matrix-size image dimension with a conventionally frequency encoded but small-matrix-size image dimension. The model-based reconstruction yields images free of distortions due to gradient non-linearity and shows overall improved in vivo male pelvic image quality. |
|
4624.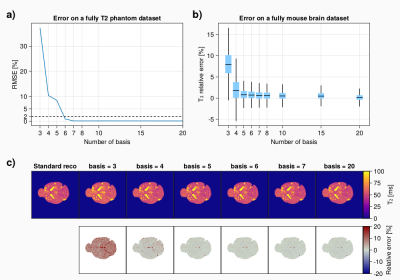 |
29 |
Accelerated 3D Multi-Echo Spin-Echo sequence for T2 mapping of
mouse brain metastases using a subspace-based reconstruction
Aurélien J. TROTIER1,
Nadège Corbin1,
Sylvain Miraux1,
and Emeline J. Ribot2
1Centre de Résonance Magnétique et Systèmes Biologiques, UMR5536, CNRS, University of Bordeaux, Bordeaux, France, 2CRMSB - University of Bordeaux / CNRS, Bordeaux, France Keywords: Pulse Sequence Design, Sparse & Low-Rank Models T2 mapping is an important biomarker for pre-clinical imaging to characterize the grows of metastasis but suffers from long acquisition time when performed in 3D. Undersampling of the k-space combined to advanced iterative reconstruction using the temporal redondancy of the data along the echo train could be beneficial to reach a reasonable scan time. This works demonstrates that a subspace-based reconstruction for T2 mapping can be used to accelerate a 3D Multi-Echo Spin-Echo acquisition for in-vivo mouse brain metastasis quantification with an acceleration factor up to 10 while keeping an isotropic spatial resolution of 156µm. |
|
4625.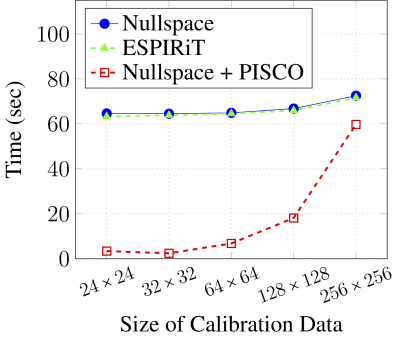 |
30 |
New Theory and Faster Computations for Subspace-Based
Sensitivity Map Estimation
Rodrigo A. Lobos1,
Chin-Cheng Chan1,
and Justin P. Haldar1
1University of Southern California, Los Angeles, CA, United States Keywords: Parallel Imaging, Sparse & Low-Rank Models Sensitivity map estimation is important in many multichannel MRI applications. Subspace-based sensitivity map estimation methods like ESPIRiT are popular and perform well, though can be computationally expensive and their theoretical principles can be nontrivial to understand. In this work, we derive a new theoretical framework for sensitivity map estimation from a structured low-rank modeling perspective. This results in an estimation approach that is equivalent to ESPIRiT, but with theory that may be more intuitive for some readers. In addition, we propose a set of computational acceleration techniques that enable substantial (~25-fold) improvements in computational time for subspace-based sensitivity map estimation. |
|
4626.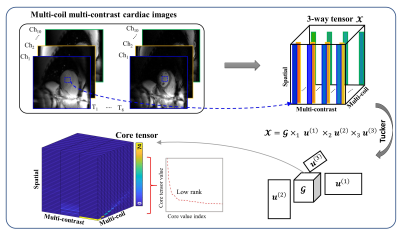 |
31 |
Accelerated Calibrationless MR Parametric Mapping by a
Space-Contrast-Coil-Domain Locally Low-Rank Tensor Constraint
Juan Gao1,
Sha Hua2,
Xin Tang1,
Haiyang Chen1,
Yixin Emu1,
and Chenxi Hu1
1Institute of Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Department of Cardiovascular Medicine, Ruijin Hospital Lu Wan Branch, Shanghai Jiao Tong University School of Medicine, Shanghai, China Keywords: Sparse & Low-Rank Models, Quantitative Imaging Parametric mapping is routinely used in cardiac MR, yet its resolution is relatively low due to the single-shot acquisition. Most existing acceleration methods exploit the space-contrast-domain and coil-domain information redundancy separately e.g. by combining LLR and SENSE. Here we propose a novel calibrationless parametric mapping acceleration technique based on a Locally Low-Rank Tensor (LLRT) modeling of the signal in the space-contrast-coil domain, which exploits the information redundancy over all 3 dimensions jointly. In vivo studies show that the method generates more accurate reconstructions than the LLR-based algorithm. Moreover, a nonuniform LLRT penalty further improves the reconstruction quality by reducing blurring. |
|
4627.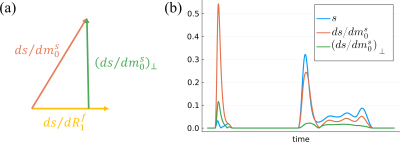 |
32 |
Cramér-Rao Bound Optimized Linear Bases for Low-Rank Subspace
Reconstruction
Andrew Mao1,2,3,
Sebastian Flassbeck1,2,
Cem Gultekin4,
and Jakob Asslaender1,2
1Center for Biomedical Imaging, New York University Grossman School of Medicine, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research, New York University Grossman School of Medicine, New York, NY, United States, 3Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine, New York, NY, United States, 4Courant Institute of Mathematical Sciences, New York University, New York, NY, United States Keywords: Sparse & Low-Rank Models, Magnetization transfer, MR Fingerprinting, Hybrid State, Cramer-Rao bound, Quantitative Imaging, Low-Rank Reconstruction This works extends the traditional framework for estimating low-rank bases that maximize preserved signal energy to additionally preserve the Cramér-Rao bound of the biophysical parameters in quantitative imaging. To this end, we orthogonalize the signal's derivatives wrt. the model parameters and incorporate them into the basis estimation process. We demonstrate in silico an improvement in the Cramér-Rao bound of all biophysical parameters with negligible cost to signal energy preservation, which translates to improved image quality and SNR in vivo. |
|
4628.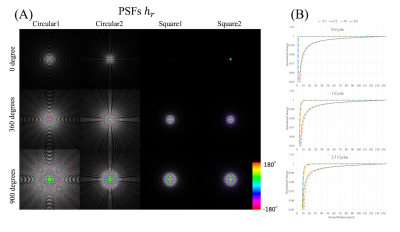 |
33 |
Improving Spiral Deblurring with Square Kernels and Low-Pass
Preconditioning
Dinghui Wang1,
Tzu Cheng Chao1,
and James G Pipe1
1Department of Radiology, Mayo Clinic, Rochester, MN, United States Keywords: Image Reconstruction, Artifacts, spiral imaging, deblurring, off-resonance Advantages of spiral imaging include fast scan speed and high SNR efficiency. Success implementation of spiral imaging relies on utilization of efficient deblurring methods. The goal of this work is to improve the performance of a previous deblurring method, using square kernels and low-pass preconditioning. Data from phantom and volunteers demonstrate that artifacts can be reduced while computational demand is also reduced by the proposed kernels.
|
|
4629. |
34 |
The Problem of Hidden Noise in MR Image Reconstruction
Jiayang Wang1,
Di An1,
and Justin Haldar1
1Signal and Image Processing Institute, Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence The performance of modern image reconstruction methods is commonly judged using quantitative error metrics like mean squared-error and the structural similarity index, where these error metrics are calculated by comparing a reconstruction against fully-sampled reference data. In practice, this reference data contains noise and is not a true gold standard. In this work, we demonstrate that this “hidden noise” can confound performance assessment methods, leading to image quality degradations when typical error metrics are used to tune image reconstruction performance. We also demonstrate that a new error metric, based on the non-central chi distribution, helps resolve this issue. |
|
4630.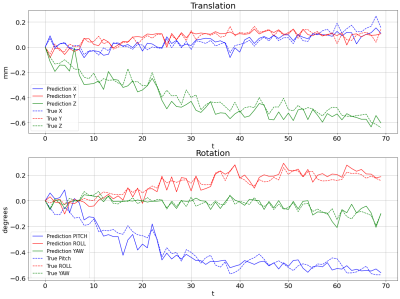 |
35 |
K-space Based Motion Estimation for polar fMRI using Transfer
learning
Faeze makhsousi1,
Vahid Ghodrati2,
Morteza Homayounfar1,
sina ghaffarzadeh1,
and abbas Nasiraei-Moghaddam3
1Biomedical Engineering, Amirkabir University of Technology (Tehran Polytechnic), Tehran, Iran (Islamic Republic of), 2University of California, Los Angeles, Los Angeles, CA, United States, 3Amirkabir University of Technology (Tehran Polytechnic), Tehran, Iran (Islamic Republic of) Keywords: Motion Correction, Brain, Data Analysis The motion of the head during functional MRI is an unavoidable issue that adversely affects brain mapping. Radial reading of the k-space reduces the problem to some extent but not completely. Residual motion, even at a partial pixel level, has a measurable effect on the spatial frequencies and so can be estimated directly from the k-space data. This work uses a transfer learning-based approach to estimate the head motion from radially acquired k-space information. Results showed a good agreement with the statistical parametric mapping (SPM) package. |
|
4631.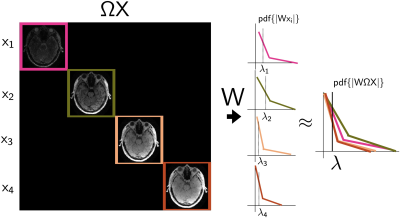 |
36 |
Concatenated multi-contrast wavelet-based compressed sensing
reconstruction.
Gabriel Varela-Mattatall1,2,
Jaejin Cho3,4,
Omer Oran5,
Corey A. Baron1,2,
Berkin Bilgic3,4,
and Ravi S. Menon1,2
1Centre for Functional and Metabolic Mapping (CFMM), Robarts Research Institute, Western University, London, ON, Canada, 2Department of Medical Biophysics, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, 3Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital & Harvard Medical School, Boston, MA, United States, 4Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Boston, MA, United States, 5Siemens Healthcare Limited, Oakville, ON, Canada Keywords: Image Reconstruction, Image Reconstruction There is a family of multi-contrast sequences such as MEGRE, MP2RAGE and 3D-QALAS which produce a limited number of contrasts, and their corresponding reconstructions in highly accelerated acquisitions are not optimally represented by either compressed sensing, low rank, or both reconstruction styles together. In this work, we explore a novel way to perform compressed sensing for simultaneous multi-contrast reconstruction. By using a concatenation operator in the regularization term, we can perform a single compressed sensing reconstruction with automatic parameter selection to reconstruct all contrasts at once and provide an implicit form of parallelization for multi-contrast reconstruction. |
|
4632.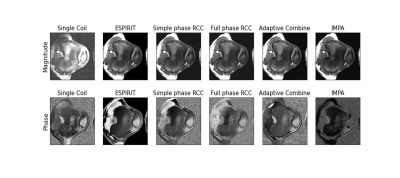 |
37 |
Comparison of coil combination technique performance for phase
preservation
Krithika Balaji1,
Peter J Lally2,3,
Zimu Huo4,
Michael Mendoza4,
Michael N Hoff5,
and Neal K Bangerter4
1Bioengineering, Imperial College London, London, United Kingdom, 2Department of Brain Sciences, Imperial College London, London, United Kingdom, 3UK Dementia Research Institute Centre for Care Research and Technology, London, United Kingdom, 4Department of Bioengineering, Imperial College London, London, United Kingdom, 5Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States Keywords: Data Processing, Cartilage, Comparisons, bSSFP MRI images are typically acquired using multiple receive coils, which introduce spatially varying phase offsets to each coil image. Many techniques have been developed to combine these coil images. For a variety of reconstruction techniques, phase preservation is necessary post-combination but it is unclear which method best achieves this. This work compared ESPIRiT, Simple Phase RCC, Full Phase RCC, 3T Siemens’ Adaptive Combine and IMPA using phase-cycled bSSFP images. Phase preservation was evaluated using the elliptical signal model theory and characteristic bSSFP phase plots. ESPIRiT consistently produced combined images with phase characteristics most similar to those from a single coil. |
|
4633.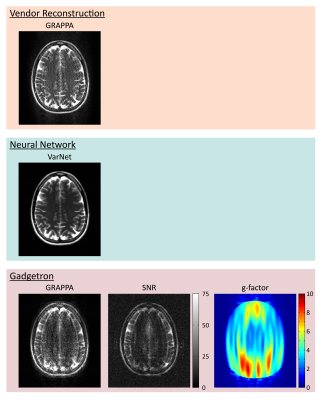 |
38 |
Standardization of Containerized “MRD Apps” for Reproducible and
Deployable Research
Kelvin Chow1,
Tess Wallace2,
Alexander Fyrdahl3,
Peter Kellman4,
Hui Xue4,
Florian Knoll5,
and Adrienne E Campbell-Washburn4
1Siemens Medical Solutions USA, Inc., Chicago, IL, United States, 2Boston Children's Hospital, Boston, MA, United States, 3Karolinska Institutet, Karolinska, Sweden, 4National Institutes of Health, Bethesda, MD, United States, 5Friedrich-Alexander Universität, Erlangen, Germany Keywords: Software Tools, Reproductive Reproducibility and translation of advancements in MR image reconstruction and analysis algorithms have been limited by incompatibilities between heterogeneous software environments. Containerization of “MRD Apps” packages software together with all dependent libraries into a single ready-to-use container. Standardization of the container format enables broad inter-compatibility and source code examples are provided for Python, MATLAB, and C++ through Gadgetron. A neural network was implemented in the MRD Apps format using Python and run directly on the scanner with vendor-provided integration. MRD Apps can be published alongside manuscripts as reference implementations or used to streamline image reconstruction and analysis challenge contests. |
|
4634.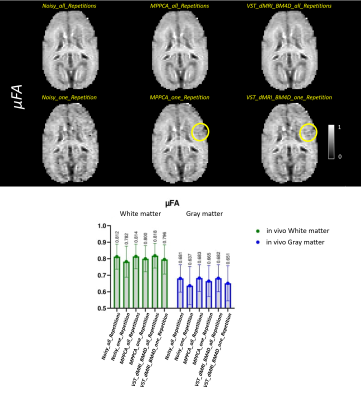 |
39 |
Exploiting diffusion MRI data redundancy on a denoising
framework - application on MR images (OGSE, μFA protocols) of
the Marmoset Brain (in-vivo)
Vinicius P. Campos1,
Tales Santini2,
Corey Baron2,
and Marcelo A. C. Vieira1
1Computer and Electrical Engineering, University of São Paulo, São Carlos, Brazil, 2Robarts Research Institute, Western University, London, ON, Canada Keywords: Data Processing, Data Analysis, Denoise Oscillating gradients spin-echo (OGSE) and microscopic fractional anisotropy (μFA) are diffusion MRI advanced techniques able to provide additional information of the microstructures of the brain1-5, when compared to traditional diffusion MRI. However, high-resolution DWI images present low signal to noise ratio (SNR). In this work, we presented a different framework, named VST_dMRI_BM4D, for denoising dMRI data by exploiting data redundancy and using the variance stabilization transformation (VST) concept7-9. Results show the proposed method is comparable to, and in some cases superior than, MPPCA6, potentially making it a useful tool to be used in the dMRI field. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.