Digital Poster
AI-Enhanced Processing in the Brain
ISMRM & ISMRT Annual Meeting & Exhibition • 04-09 May 2024 • Singapore

| Computer # | |||
|---|---|---|---|
4512.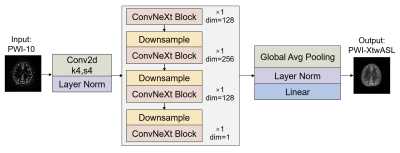 |
33 |
Denoising Arterial Spin Labeling Perfusion MRI using Deep
Learning Methods
Zixuan Han1,
Zihan Ning1,
and Xihai Zhao1
1Center for Biomedical Imaging Research, Tsinghua University, Beijing, China Keywords: Other AI/ML, Machine Learning/Artificial Intelligence, Arterial Spin Labeling Motivation: The low SNR of arterial spin labeling (ASL) perfusion weighted images (PWI) directly affects the accuracy of cerebral blood flow (CBF) quantification. Goal(s): Our goal was to improve SNR of ASL PWI and access accurate CBF quantification. Approach: We developed XtwASL, a deep learning model for denoising ASL PWI. Evaluation included SNR, peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM) for PWI quality as well as ICC and Bland-Altman analysis for CBF assessment. Results: Compared with other methods, the proposed XtwASL significantly improved SNR of ASL in the case of short acquisition time and the CBF after XtwASL denoising was more accurate. Impact: The ASL denoising method XtwASL not only improves SNR but also has good clinical usability in CBF quantification, and this may reduce patient discomfort and artifacts caused by long scan time especially in multi-PLD ASL and high-resolution ASL field. |
|
4513.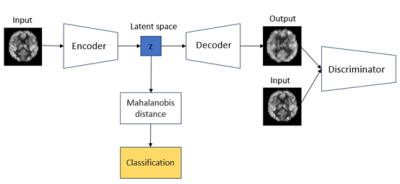 |
34 |
Automated Quality Control for Arterial Spin Labelling (ASL) MRI
Using a VAE-based Neural Network
Jian Hu1,2,
Silvin P. Knight3,
Bowen Deng4,
Rose Anne Kenny3,5,6,
Xin Chen7,
and Michael Chappell1,2
1Mental Health & Clinical Neurosciences, School of Medicine, University of Nottingham, Nottingham, United Kingdom, 2Sir Peter Mansfield Imaging Centre, School of Medicine, University of Nottingham, Nottingham, United Kingdom, 3The Irish Longitudinal Study on Ageing (TILDA), School of Medicine, Trinity College Dublin, Dublin, Ireland, 4Computer Vision Laboratory, School of Computer Science, University of Nottingham, Nottingham, United Kingdom, 5Discipline of Medical Gerontology, School of Medicine, Trinity College Dublin, Dublin, Ireland, 6Mercer’s Institute for Successful Ageing (MISA), St. James’s Hospital, Dublin, Ireland, 7Intelligent Modelling&Analysis Group, School of Computer Science, University of Nottingham, Nottingham, United Kingdom Keywords: Diagnosis/Prediction, Perfusion, Quality Control Motivation: Quality Control (QC) of ASL data is primarily a manual and subjective process that is time-intensive and can yield inconsistent results due to rater variability, while existing tools provide limited diagnostic metrics mostly focused on specific error source. Goal(s): To develop a QC detector to automatically detect outliers for ASL data. Approach: VAE-GAN model was applied to extract the latent representation of ASL data by which the decision boundary can be determined. Results: The AUROC of our QC detector on test dataset is 0.82 with accuracy=0.81. Impact: Our QC detector could help radiologists and researchers working on ASL MRI to automatically identify outliers. Consequently, appropriate operations can be used to correct or exclude outliers to avoid biases in the outcomes and ensure accurate interpretations. |
|
4514.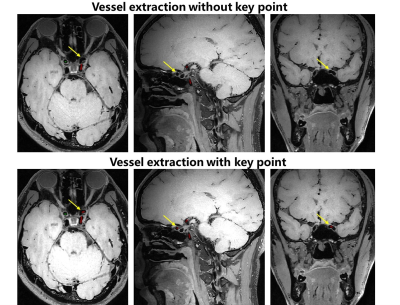 |
35 |
Deep Learning Based Key Point Detection Algorithm to Assist
Accurate Vascular Centerline Extraction and Automatic
Quantitative Plaque Analysis
Xiqian Zhang1,2,
Wanqing Sun3,
Xiong Yang3,
Yufei Mao3,
Ye Li1,2,4,5,
Dong Liang1,2,4,5,
Xin Liu1,2,4,5,
Hairong Zheng1,2,4,5,
and Na Zhang1,2,4,5
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Department of Image Advanced Analysis of HSW BU, Shanghai United Imaging Healthcare Co., Ltd., Shanghai, China, 4Key Laboratory of Biomedical Imaging Science and System, Chinese Academy of Sciences, Shenzhen, China, 5United Imaging Research Institute of Innovative Medical Equipment, Shenzhen, China Keywords: Diagnosis/Prediction, Machine Learning/Artificial Intelligence, Plaque analysis;Vascular centerline extraction Motivation: Magnetic Resonance Vessel Wall Imaging is a crucial method for assessing arterial plaques. Goal(s): Achieving a rapid and accurate algorithm for vascular centerline extraction and automatic detection of the pituitary stalk, thereby enabling fast, automatic, and quantitative analysis of plaques. Approach: Proposed a point detection algorithm based on V-NET, designed to achieve rapid and accurate extraction of vascular centerlines and automatic localization of the pituitary stalk. Results: Accurate key point detection had been achieved, enhancing the precision of vascular centerline extraction and enabling fast and accurate automatic localization of the pituitary stalk. Impact: The accuracy of vascular centerline detection has been improved, and rapid and automatic quantitative analysis of plaque enhancement has been realized. This can assist in enhancing diagnostic efficiency in clinical settings. |
|
4515.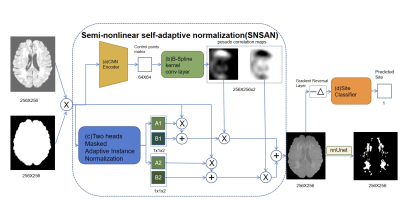 |
36 |
Semi-Nonlinear Self-Adaptation Normalization Generator (SNSAN)
to Improve Generalization of White Matter Hyperintensities
Segmentation
Yu Cheng1,
Chengyan Wang2,
Beini Fei3,
Chun-Yi Zac Lo1,4,
and He Wang1
1The Institute of Science and Technology for Brain-inspired Intelligence (ISTBI), Fudan University, Shanghai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Zhongshan Hospital Fudan University, Shanghai, China, 4Department of Biomedical Engineering, Chung Yuan Christian University, Taoyuan, Taiwan Keywords: AI Diffusion Models, White Matter, WMH segmentation, Domain Generalization, Normalization Motivation: Self-adaptation normalization (SAN) method generalizes lesion segmentation to new sites by using a linear generator to convert inputs to a site-independent style. But it fails when the domain difference is mostly nonlinear. Goal(s): Develop a semi-nonlinear self-adaptation network(SNSAN) to generalize the White Matter Hyperintensity(WMH) segmentation model to an external site. Approach: To replace the linear generator, the method blends two SAN results with a pseudo correlation map, later use the gradient reversal method to guide the result to a site-unrelated style. Results: SNSAN normalizes the input data close to a Gaussian distribution and improves the generalization performance on the data from external site. Impact: We provide a simple and efficient semi-nonlinear normalization method to enhance the domain generalization, and its performance is better than SAN when the domain gap is affected by more nonlinear factors. |
|
4516.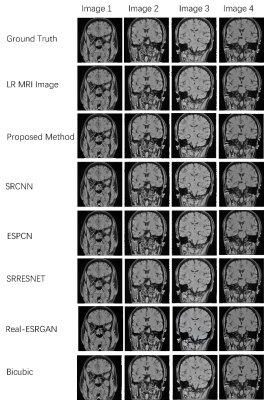 |
37 |
ResoNet: MRI Super-Resolution via Multi-Path GAN with Adaptive
Convolutional Kernels
Haotian Zhang1,
Long Teng1,
Yi Zhao2,
Hang Qu2,
Weiqiang Dou3,
Qi-qi Ban2,
and Gangzhong Wen4
1The Hong Kong Polytechnic University, HongKong, China, 2Affiliated Hospital of Yangzhou University, Yangzhou, China, 3GE Healthcare, MR Research China, Beijing, China, 4Shenzhen Wrmherd biotech Ltd. co., Shenzhen, China Keywords: Diagnosis/Prediction, Vessels, Image super-resolution , Generative Adversarial Network (GAN) , Multi- path generator model , Medical imaging Motivation: The project aims to improve MRI image resolution, addressing the limitations of current super-resolution methods that hinder accurate medical diagnosis and treatment planning. Goal(s): Enhancing MRI image clarity through ResoNet, an innovative approach featuring a multi-path generator, self-adaptive kernel sizing, and a comprehensive loss function. Approach: Multi-path generator enhances resolution with different kernel sizes, preserving details. Self-adaptive component optimizes kernel size for efficiency. Comprehensive loss function combines GAN-based discriminator, VGG16, and WGAN-GP for sTab. training and quality samples. Results: ResoNet significantly enhances SSIM and PSNR scores, promising substantial impact in medical imaging for more precise diagnoses and treatment planning by healthcare professionals. Impact: This work improves MRI diagnostics by enhancing image resolution, enabling clearer visuals for accurate medical assessments. It optimizes performance and resources, accelerates scanning time, and potentially transforms emergency medical practices, making it an improvement in medical imaging. |
|
4517.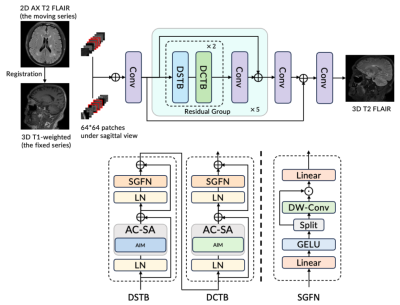 |
38 |
Generating 3D Volumetric Brain Images via Other Contrast
Guidance from 2D Thick Slices
Long Wang1,
Lei Xiang1,
Ryan Chamberlain1,
Xinyu Song2,
Xiao-Er Wei2,
and Yuehua Li2
1Subtle Medical, Menlo Park, CA, United States, 2Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China Keywords: AI/ML Image Reconstruction, Multi-Contrast
Motivation: The conventional multiple sclerosis(MS)
protocol in the brain normally takes from 30 minutes to
several hours. It often involves multiple 3D series for the
quantitative metrics and reports. Goal(s): To reduce the scan time for the brain MS protocols with more than one 3D series Approach: We proposed a framework that utilized a 3D T1 and 2D FLAIR as inputs to generate a 3D T2 FLAIR.
Results: The method generates comparable image
resolution on all orientations as the acquired 3D FLAIR, and
outperforms other methods. The lesion segmentation masks
show high consistency. Furthermore, the method demonstrates
robustness in the cases with motion. Impact: The method provides the solution of generating a volumetric FLAIR using the same time as a 2D series and achieving comparable resolutions and diagnosis accuracy as the 3D FLAIR. |
|
4518.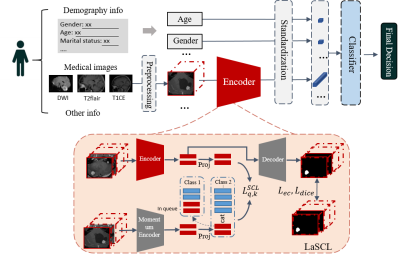 |
39 |
A Computer-aided Diagnoses Method for Brainstem-and-cerebellar
Hemangioblastoma
Botao Zhao1 and
Xiao-Yong Zhang1
1Fudan University, Shanghai, China Keywords: Diagnosis/Prediction, Tumor, Hemangioblastoma, Contrastive Learning Motivation: Brainstem and cerebellar hemangioblastoma is a rare tumor with a high risk of haemorrhage during the biopsy. However, it is still a challenging task to distinguish HB from other types of intracranial tumors solely based on neuroimaging techniques. Goal(s): To propose a computer-aided diagnosis method to classify hemangioblastoma. Approach: We propose a patient-level classification framework using multi-task supervised contrastive learning, named LaSCL-PLC, for hemangioblastoma classification. Results: We evaluated the proposed model on a local MRI dataset of brainstem-and-cerebellum tumors, consisting of 97(HB) and 143(others). The experimental results show that our model achieves competitive performance as neuroradiologists. Impact: It could improve the preoperative diagnosis hemangioblastoma, which is crucial for clinical treatments. |
|
4519.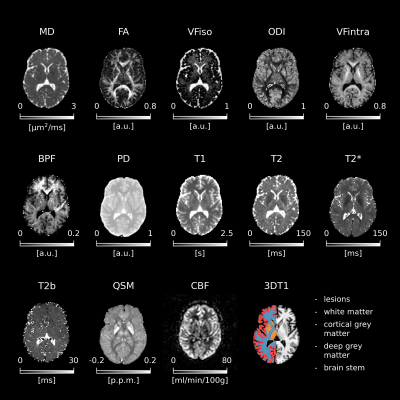 |
40 |
Random forest classifiers show myelin alterations in Long-COVID
Antonio Ricciardi1,
Elena Grosso2,
Madiha Shatila1,
Michael S. Zandi3,
Marios C. Yiannakas1,
Ferran Prados1,4,5,
Baris Kanber1,4,
Jed Wingrove1,
Francesco Grussu1,6,
Marco Battiston1,
Rebecca S. Samson1,
Carmen Tur7,
Rachel L. Batterham8,9,
Janine Makaronidis8,9,
Nicolò Rolandi1,2,10,
Fulvia Palesi2,11,
Egidio D'Angelo2,11,
Olga Ciccarelli1,9,
and Claudia A. M. Gandini Wheeler-Kingshott1,2,11
1NMR Research Unit, Queen Square MS Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 2Department of Brain & Behavioural Sciences, University of Pavia, Pavia, Italy, 3Department of Brain Repair and Rehabilitation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 4Department of Medical Physics and Biomedical Engineering, Centre for Medical Image Computing (CMIC), University College London, London, United Kingdom, 5E-Health Center, Universitat Oberta de Catalunya, Barcelona, Spain, 6Radiomics Group, Vall d’Hebron Institute of Oncology, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain, 7Neurology-Neuroimmunology Department Multiple Sclerosis Centre of Catalonia (Cemcat), Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain, 8Centre for Obesity Research, Department of Medicine, University College London, London, United Kingdom, 9National Institute of Health Research, Biomedical Research Centre at UCLH and UCL, University College London, London, United Kingdom, 10Department of Clinical and Experimental Epilepsy, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 11Digital Neuroscience Center, IRCCS Mondino Foundation, Pavia, Italy Keywords: Analysis/Processing, COVID-19, Long-COVID, Multimodal, MRI, myelin.
Motivation: Some people exposed to SARS-CoV-2
experience persistent symptoms months after recovering from
acute infection. These symptoms, grouped under the umbrella
of “long-COVID”, include neurological and autonomic
dysfunctions for which there is not yet a clear explanation.
Goal(s): To identify biophysically meaningful
features characterising long-COVID pathophysiology in the
brain.
Approach: A novel multimodal MRI protocol was
developed to measure iron accumulation, myelin content,
inflammation, microstructure alterations, atrophy, and
cerebral blood flow changes. Machine learning was employed
to discriminate long-COVID from controls and people who
fully recovered.
Results: Alterations in bound-pool fraction (myelin
content) emerge in long-COVID.
Impact: This study encourages investigating the
neurological aspect of long-COVID and its interplay with
conditions affecting myelin as it shows that long-COVID may
cause myelin alterations or be more likely to occur in
people with compromised myelin content in the brain. |
|
4520.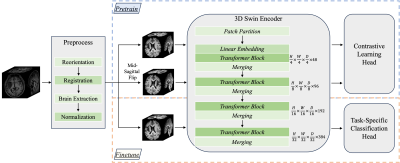 |
41 |
Asymmetry-Aware Pretraining: A Path to Universal Brain MRI
Analysis Model
Yang Ma1,2,
Dongang Wang2,3,
Peilin Liu2,4,
Michael Barnett2,3,5,
Weidong Cai1,
and Chenyu Wang2,3
1School of Computer Science, University of Sydney, Sydney, Australia, 2Brain and Mind Centre, University of Sydney, Sydney, Australia, 3Sydney Neuroimaging Analysis Centre, Sydney, Australia, 4School of Mathematics and Statistics, University of Sydney, Sydney, Australia, 5Department of Neurology, Royal Prince Alfred Hospital, Sydney, Australia Keywords: Diagnosis/Prediction, Brain
Motivation: Many brain disorders present with
asymmetrical features, which can be utilized to advance the
classification of disease through deep learning. Goal(s): We aim to investigate how asymmetry awareness can improve model performance to identify brain disorders, subsequently facilitating better early diagnosis and intervention.
Approach: Our approach involved initial pretraining
with a focus on asymmetry awareness using conditional
contrastive learning. Subsequently, finetuning was performed
for various downstream tasks with limited training data.
Results: Our approach is proven superior in
identifying brain disorders compared with baseline methods
that either directly trained for specific diseases or with
pretrained model without considering symmetrical features of
input images.
Impact: Our research pioneers the integration of
asymmetry awareness in pretraining models for the detection
of brain disorders. The superior performance fully
illustrated the feasibility and potential of leveraging the
asymmetry nature of brain disease for broad deep learning
tasks. |
|
4521.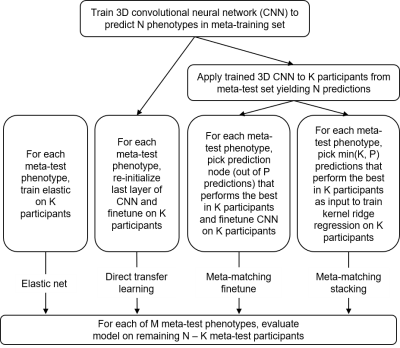 |
42 |
Meta-matching to translate phenotypic predictive models from big
to small data on structural MRI
Naren Wulan1,2,3,
Lijun An1,2,3,
Chen Zhang1,2,3,
Ru Kong1,2,3,
Pansheng Chen1,2,3,
Danilo Bzdok4,5,
Simon Eickhoff6,7,
Avram Holmes8,
and B. T. Thomas Yeo1,2,3,9,10
1Centre for Sleep & Cognition & Centre for Translational Magnetic Resonance Research, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore, 2Department of Electrical and Computer Engineering, National University of Singapore, Singapore, Singapore, 3N.1 Institute for Health & Institute for Digital Medicine, National University of Singapore, Singapore, Singapore, 4Department of Biomedical Engineering,McConnell Brain Imaging Centre (BIC), Montreal Neurological Institute (MNI), Faculty of Medicine, School of Computer Science, McGill University, Montreal, QC, Canada, 5Mila – Quebec Artificial Intelligence Institute, Montreal, QC, Canada, 6Institute for Systems Neuroscience, Medical Faculty, Heinrich-Heine University Düsseldorf, Düsseldorf, Germany, 7Institute of Neuroscience and Medicine, Brain & Behavior (INM-7), Research Center Jülich, Jülich, Germany, 8Department of Psychiatry, Brain Health Institute, Rutgers University, Piscataway, NJ, United States, 9Integrative Sciences and Engineering Programme (ISEP), National University of Singapore, Singapore, Singapore, 10Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States Keywords: Diagnosis/Prediction, Brain, Phenotypic prediction, structural MRI, transfer learning Motivation: Small sample size on structural MRI is evitable in reality and significantly limits phenotypic prediction performance. Goal(s): Our goal was to improve prediction performance on small datasets for structural MRI brain imaging. Approach: We adapted the meta-matching framework from functional to structural MRI, and compared it with baseline methods (Elastic net and direct transfer learning). Results: Our meta-matching-based approaches can greatly boost behavioral prediction performance for different small-scale structural MRI datasets. Impact: Our meta-matching-based methods should be able to make good predictions for a variety of neurological and psychiatric disorders even if the availability of structural MRI brain imaging is quite small. |
|
4522.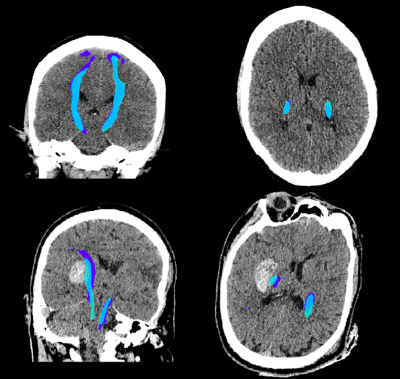 |
43 |
Deep learning to predict DTI tractography labels from diagnostic
CT scans in acute ICH
Olivia Murray1,
Hamied Haroon2,
Patel Hiren2,
George Harsten3,
Ulrike Hammerbeck4,
Marieke Wermer5,
Wilmar Jolink6,
Daniel Hanley7,
MISTIE III investigators7,
Timothy Cootes2,
Karin Klijn8,
and Adrian Parry-Jones2
1Division of Informatics, Imaging and Data Science, University of Manchester, Manchester, United Kingdom, 2University of Manchester, Manchester, United Kingdom, 3Brainomix, Oxford, United Kingdom, 4King's College London, London, United Kingdom, 5University Medical Center Groningen, Groningen, Netherlands, 6Isala Klinieken, Zwolle, Netherlands, 7Johns Hopkins, Baltimore, MD, United States, 8Radboud University Medical Center, Nijmegen, Netherlands Keywords: Diagnosis/Prediction, Diffusion Tensor Imaging Motivation: Assessing white matter integrity may enhance intracerebral haemorrhage (ICH) surgical trial selection. However, patients often only receive CT imaging, on which delineating white matter is challenging. Goal(s): This study aimed to train a model to delineate white matter in CT scans, using paired DTI tractography maps, and test if the model predicted outcome in an external dataset. Approach: Tractography maps were generated from the DTI images. A nnU-Net model was trained on paired CT and registered tractography data, and then run on an external dataset of ICH diagnostic CT scans. Results: The model performed at 58% Dice, and significantly predicted outcome after ICH. Impact: Our model can predict tractography labels of the corticospinal tract on diagnostic CT scans without the need for DTI, allowing an enhanced prediction of outcome after intracerebral haemorrhage, and potentially leading to more informed selection of candidates for surgical trials. |
|
4523.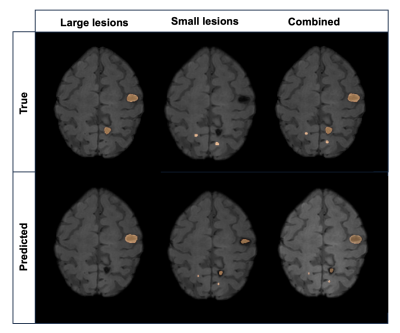 |
44 |
AUTOMATED CEREBRAL CAVERNOUS MALFORMATION LESION SEGMENTATION
FROM SUSCEPTIBILITY WEIGHTED MR IMAGING
Jinhee Jenny Lee1,2,
Marc Mabray3,
Ozan Genc1,
Jefferey Nelson4,
Helen Kim4,
and Janine M Lupo1,2
1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2UCSF/UC Berkeley Graduate Program in Bioengineering, University of California San Francisco – University of California, Berkeley, San Francisco, CA, United States, 3Department of Radiology, University of New Mexico School of Medicine, Albuquerque, NM, United States, 4Department of Anesthesia, University of California San Francisco, San Francisco, CA, United States Keywords: Analysis/Processing, Susceptibility Motivation: Familial cerebral cavernous malformation (CCM) cases typically present with multi-focal lesions of varying size, count, and location making automatic segmentation for studying lesion burden challenging. Goal(s): We aimed to develop a deep learning framework for automated detection and volumetric quantification for ongoing large-scale, multi-site evaluation of patients with CCM. Approach: We implemented a framework that ensembles two-staged deep learning networks for large and small CCM lesions. Results: Our model achieved an overall Dice-score of 70%. We demonstrated the feasibility of a deep learning approach for detecting and segmenting various-sized lesions in patients with CCM using SWI acquired from multiple sites with various parameters. Impact: Our study found that total lesion volume quantified with our deep learning approach was statistically associated with increased odds of intracranial hemorrhage history. This demonstrates the potential benefit of our approach in providing a more accurate assessment of disease burden |
|
4524.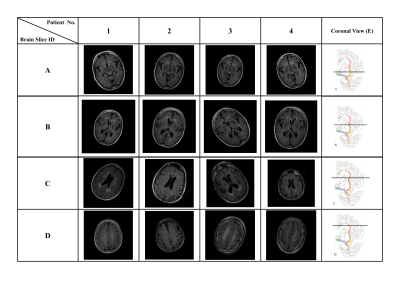 |
45 |
Using ResNet to utilize 4-class T2-FLAIR slice classification
based on the cholinergic pathways hyperintensities scale for
pathological aging
Wei-Chun Kevin Tsai1,
Yi-Chien Liu2,
Ming-Chun Yu2,
Chia-Ju Chou2,
Sui-Hing Yan3,
Yang-Teng Fan4,
Yan-Hsiang Huang3,
Yen-Ling Chiu5,
Yi-Fang Chuang6,7,
Ran-Zan Wang1,
and Yao-Chia Shih4
1Department of Computer Science and Engineering, Yuan Ze University, Taoyuan City, Taiwan, 2Department of Neurology, Cardinal Tien Hospital, New Taipei City, Taiwan, 3Department of Neurology, Far Eastern Memorial Hospital, New Taipei City, Taiwan, 4Graduate Institute of Medicine, Yuan Ze University, Taoyuan City, Taiwan, 5Department of Medical Research, Far Eastern Memorial Hospital, New Taipei City, Taiwan, 6Department of Psychiatry, Far Eastern Memorial Hospital, New Taipei City, Taiwan, 7Institute of Public Health, National Yang Ming Chiao Tung University, Taipei, Taiwan Keywords: Analysis/Processing, Machine Learning/Artificial Intelligence, T2-FLAIR, white matter hyperintensity, dementia, cholinergic pathway Motivation: Cholinergic Pathways Hyperintensities Scale (CHIPS) is a visual rating scale to evaluate the burden of cholinergic white matter hyperintensities in T2-FLAIR image, indicating the severity of dementia. However, it is still time-consuming to screen slices throughout the whole brain to choose 4 specific slices for rating. Goal(s): To develop a deep-learning-based model to automatically select 4 slices specific to CHIPS. Approach: We used ADNI T2-FLAIR dataset (N=150) to train a 4-class slice classification model (BSCA) utilized by ResNet, and a local dataset (N=30) to test its performance. Results: Our model achieved the accuracy of 99.82% and F1-score of 99.83%. Impact: BSCA can be an automatic screening tool to efficiently provide 4 specific T2-FLAIR slices covering the white matter landmarks along the cholinergic pathways for clinicians to help evaluate whether patients have the high risk to develop clinical dementia. |
|
4525.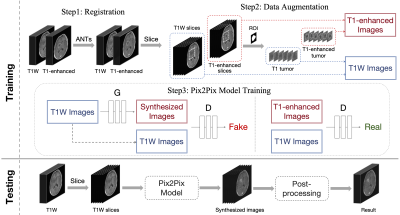 |
46 |
Brain Tumor Enhancing in Pre-contrast MRI Based on Deep Learning
Xinran Wu1,
Yaping Wu2,
Meiyun Wang2,
Rencheng Zheng1,
Weibo Chen3,
Chengyan Wang4,
and He Wang1,4
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Department of Medical Imaging, Henan Provincial People's Hospital, Zhengzhou, China, 3Philips Healthcare, Shanghai, China, 4Human Phenome Institute, Fudan University, Shanghai, China Keywords: Diagnosis/Prediction, Cancer Motivation: The project was motivated by the need to reduce reliance on contrast agents in MRI for brain tumor assessment, addressing issues of cost and patient discomfort associated with their use. Goal(s): To improve the visibility and recognition of tumors in T1-weighted MRI scans, eliminating the need for contrast agents. Approach: The approach involved registration, data augmentation, Pix2Pix-based image generation, and post-processing. Results: The results demonstrated that the method significantly improved tumor discernibility in T1-weighted MRI scans, offering a valuable diagnostic tool without the need for contrast agents. Impact: Our research provides an effective solution to enhance brain tumor visualization in MRI, directly benefiting patients and healthcare providers. By eliminating the need for contrast agents, our approach reduces costs, discomfort, and potential risks, advancing brain tumor diagnostics. |
|
4526.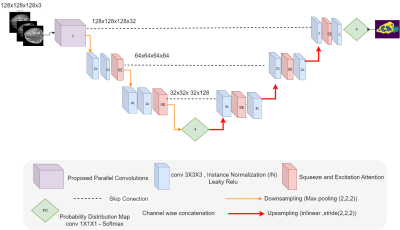 |
47 |
Assessing and Enhancing the Robustness of Brain Tumor
Segmentation using a Probabilistic Deep Learning Architecture
Ebtihal Alwadee1,2,
Xianfang Sun1,
Yipeng Qin1,
and Frank Langbein1
1Cardiff University, Cardiff, United Kingdom, 2Jazan University, Jazan, Saudi Arabia Keywords: Diagnosis/Prediction, Cancer, Analysis/Processing, Robustness
Motivation: Motivated by the challenge of enhancing
the robustness of deep neural network decisions against Goal(s): This study aims to evaluate the efficacy of probabilistic bottlenecks in enhancing segmentation robustness.
Approach: Our approach simulates structured
perturbations at increasing strength
Results: Results show probabilistic bottlenecks
significantly increase robustness to Gaussian noise, yet Impact: This study provides a tool to assess and guard against various perturbations in deep learning.It specifically demonstrates that probabilistic bottlenecks boost robustness of performance withrespect to certain noise types, but not all. |
|
4527.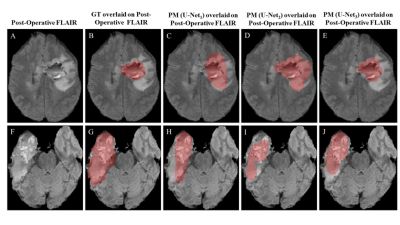 |
48 |
Post-Surgical Cavity Segmentation on MRI for Glioma Patients
with Deep Learning and Transfer Learning Approach
Virendra Kumar Yadav1,
Ankit Kandpal1,
Raufiya Jafari1,
Rakesh Kumar Singh2,
Rakesh Kumar Gupta2,
Sumeet Agarwal3,4,
and Anup Singh1,4,5
1Centre for Biomedical Engineering, Indian Institute of Technology, Delhi, New Delhi, India, 2Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 3Department of Electrical Engineering, Indian Institute of Technology, Delhi, New Delhi, India, 4Yardi School of Artificial Intelligence, Indian Institute of Technology, New Delhi, India, 5Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India Keywords: Diagnosis/Prediction, Tumor, Post cavity segmentation Motivation: Currently, manual/semiautomatic methods are being used for post-surgical cavity segmentation in glioma patients, which is crucial for precise treatment planning and monitoring. Goal(s): To evaluate the potential of deep learning models for automatic post-surgical cavity segmentation. Approach: Deep learning models were trained on pre-surgery and post-surgery (within 72hours) MRI data and tested on post-surgery data. The concept of data augmentation and transfer learning were also used. Results: Initially trained on the pre-surgery BraTS’2021 challenge dataset, the models underwent further refinement with local hospital's post-surgical data, resulting in 36% improvement in post-surgical cavity segmentation (U-Net3, DSC = 0.64). Impact: This study demonstrates the potential of deep-learning in automatic segmentation of post-surgical cavity on MRI images. Proposed automatic segmentation models can assist in fast and objective treatment planning. Further optimizations for improved accuracy and validation on large dataset is required. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.